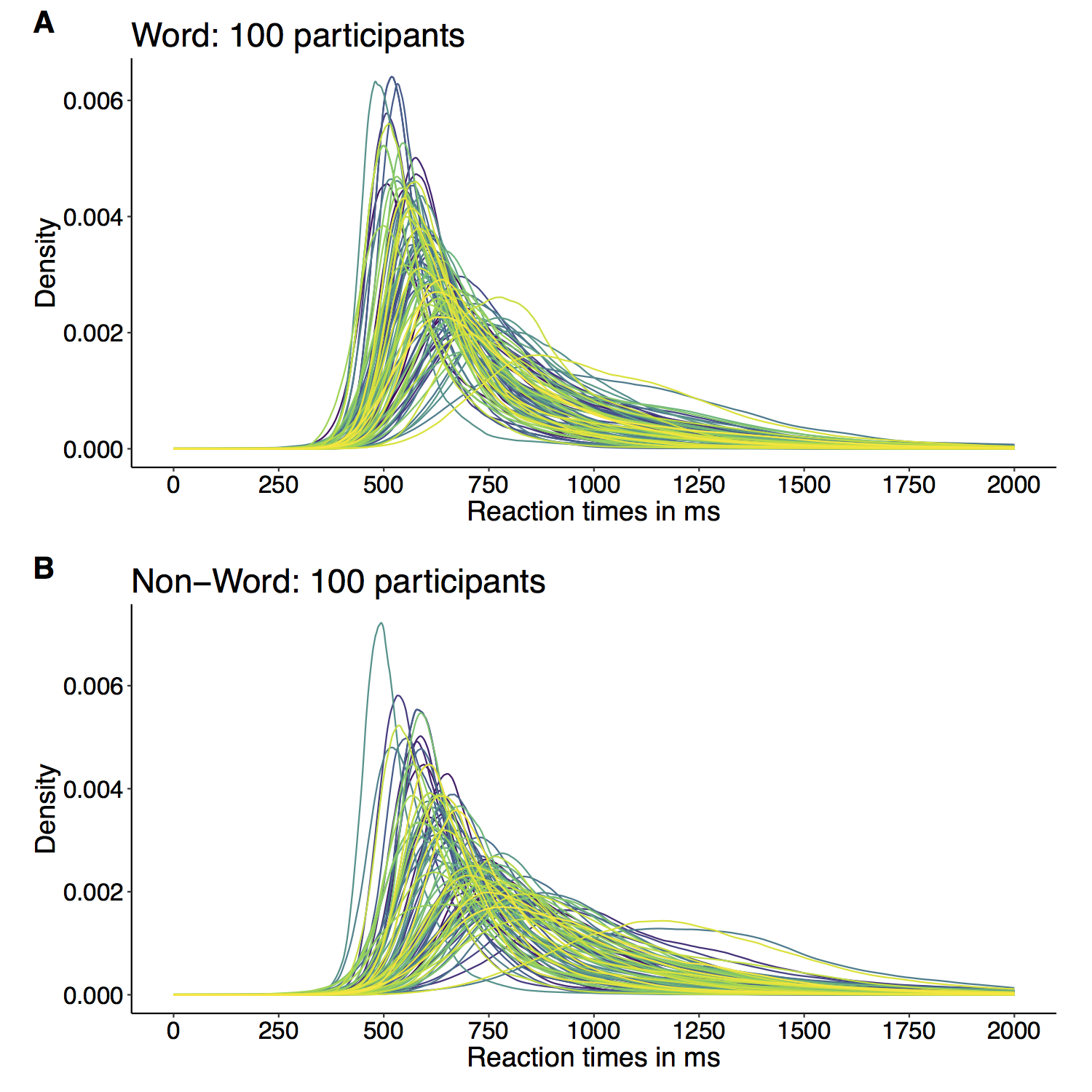
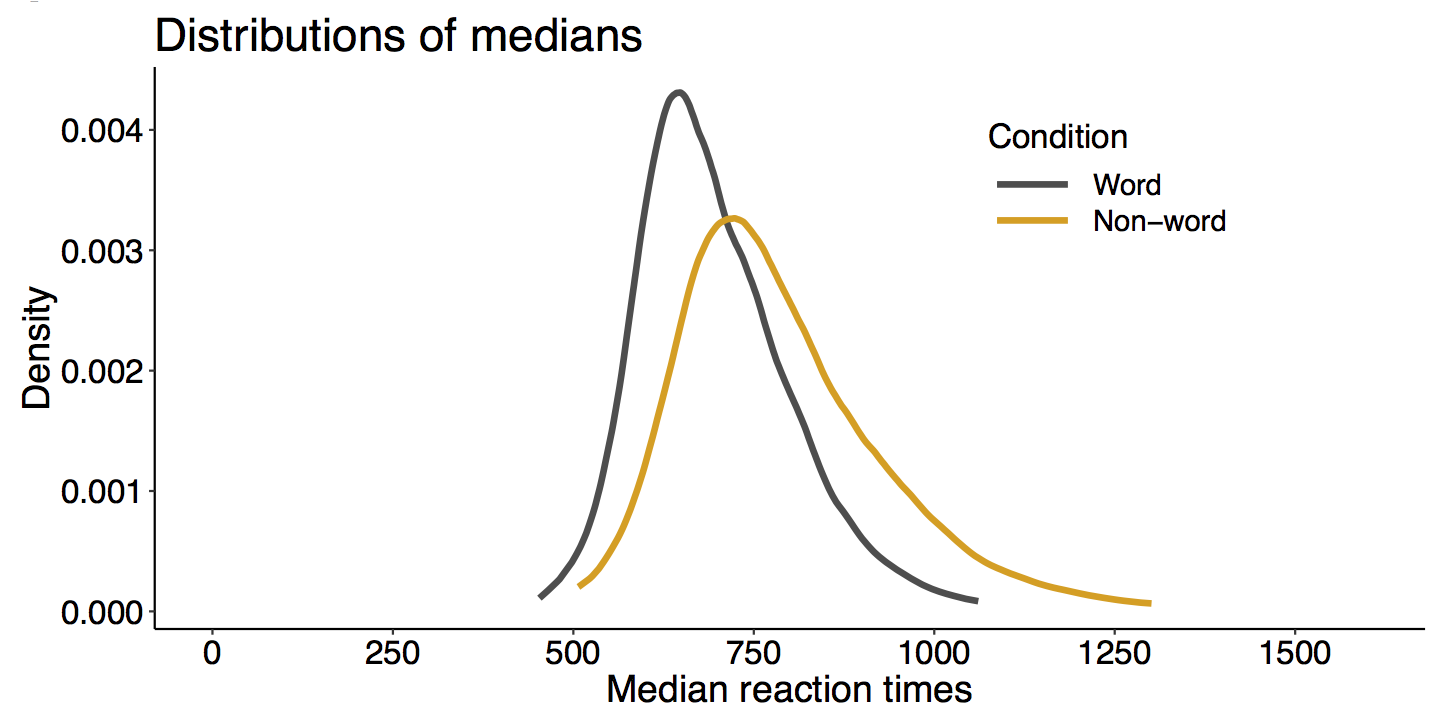
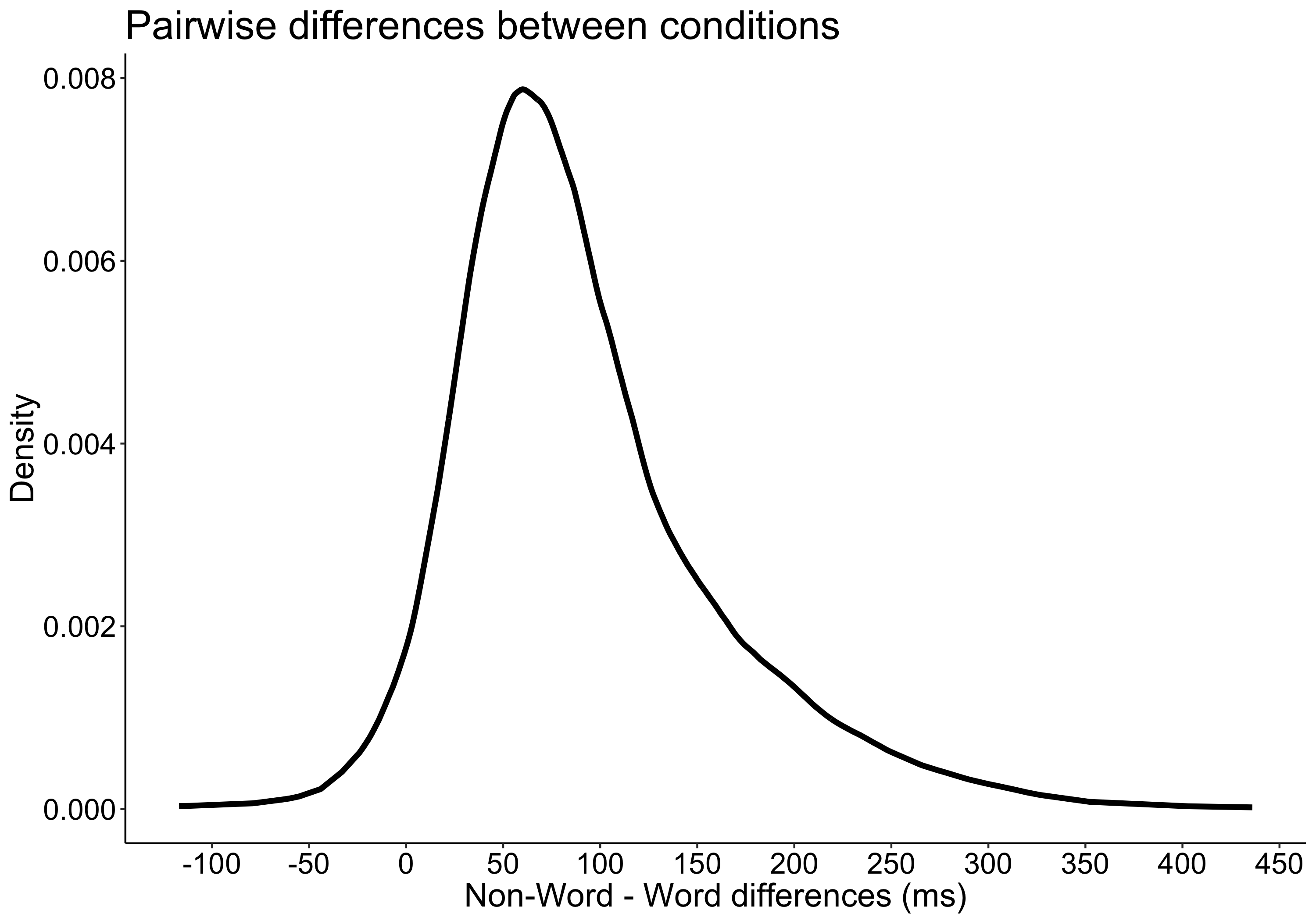
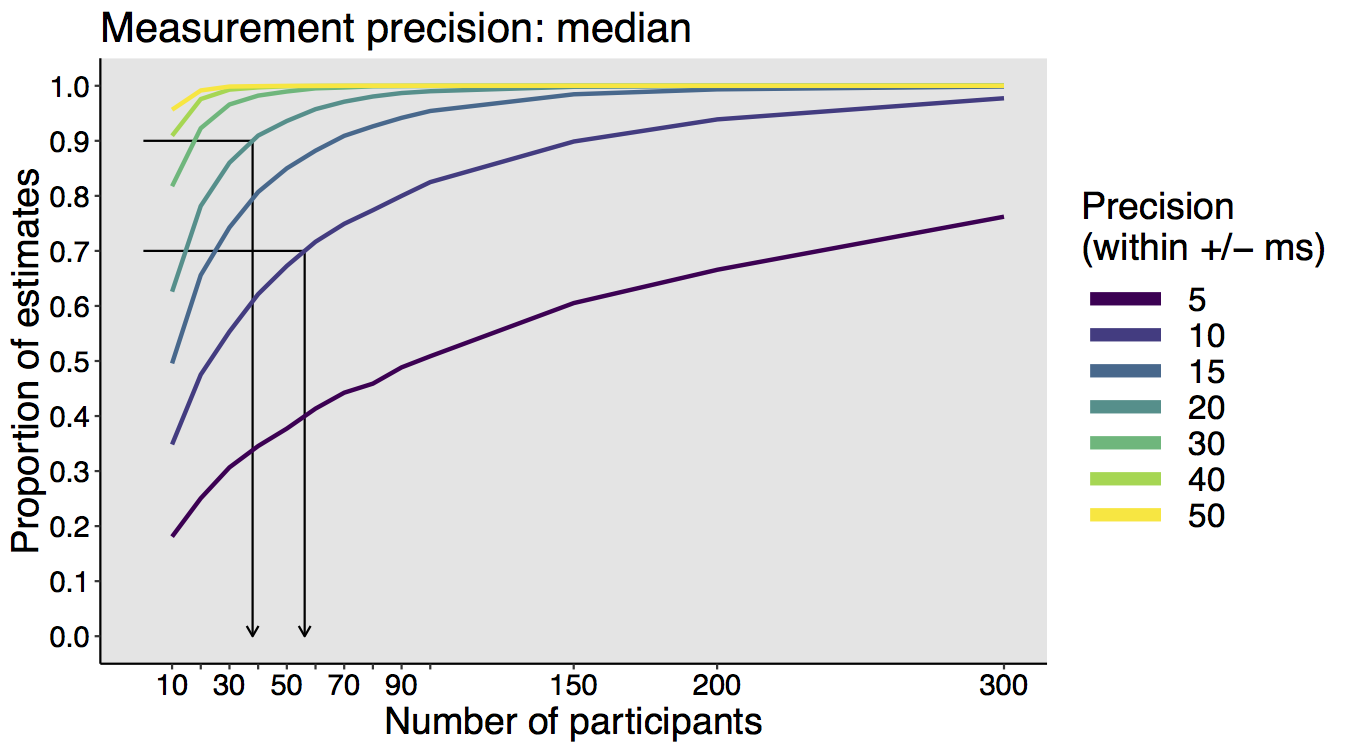
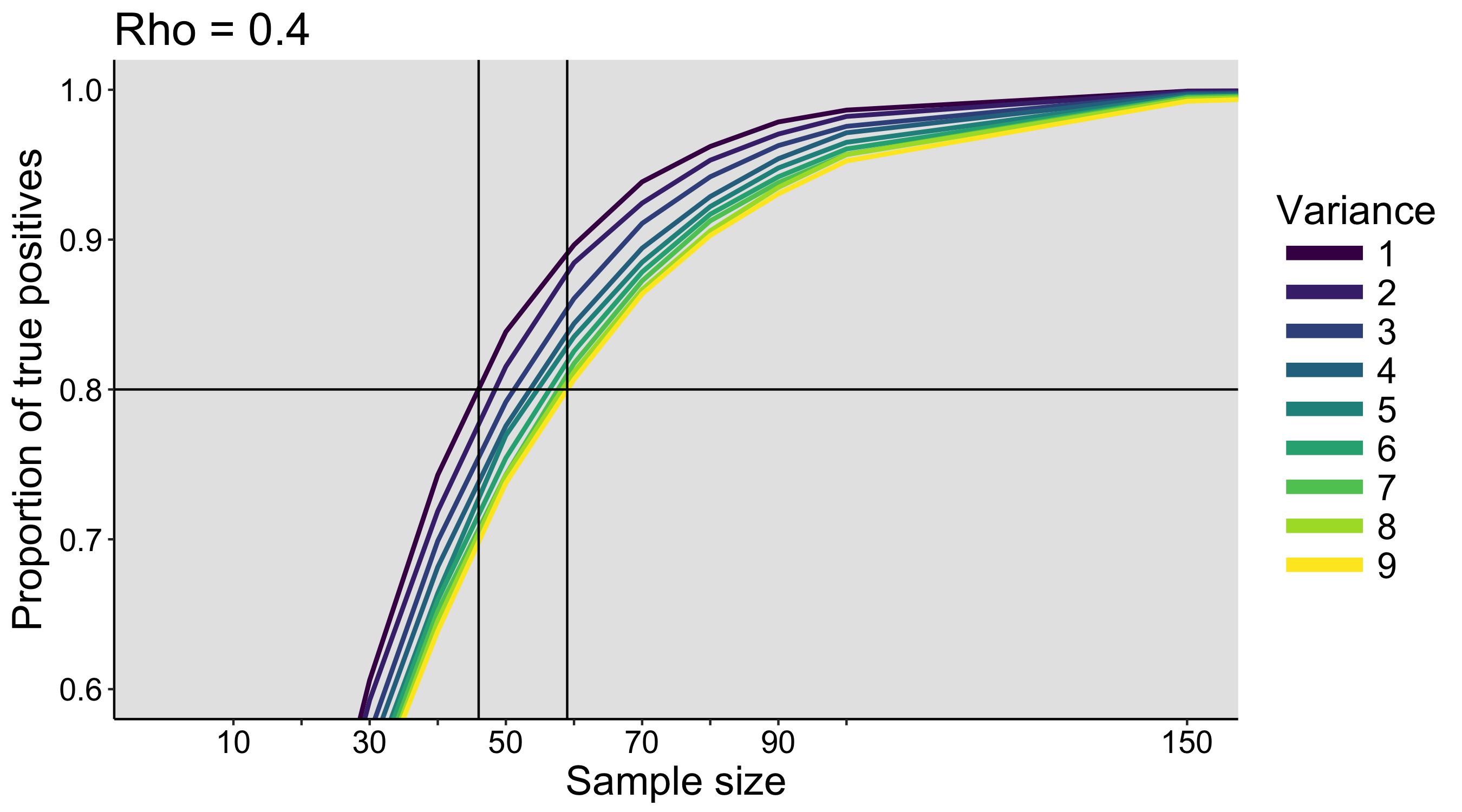
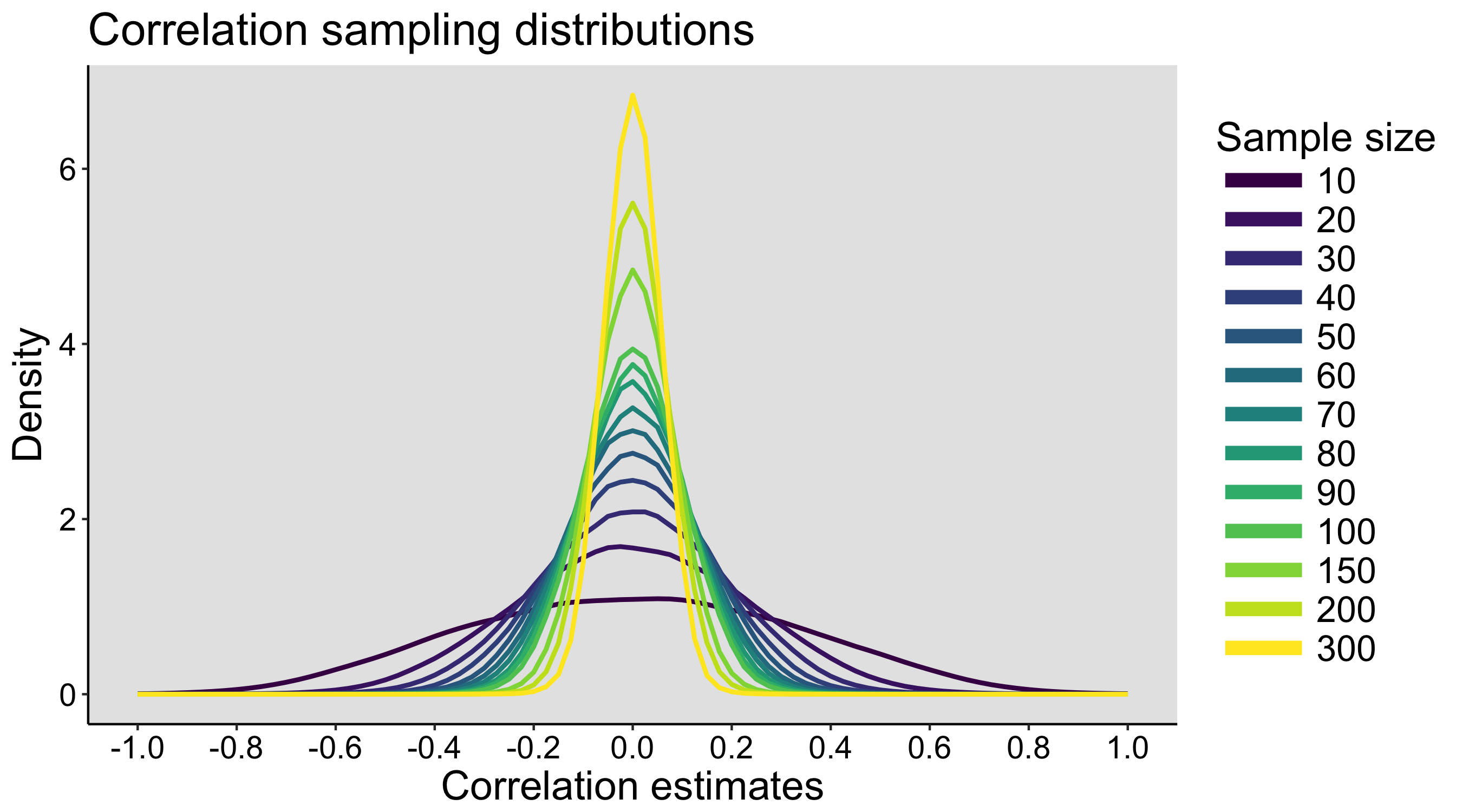
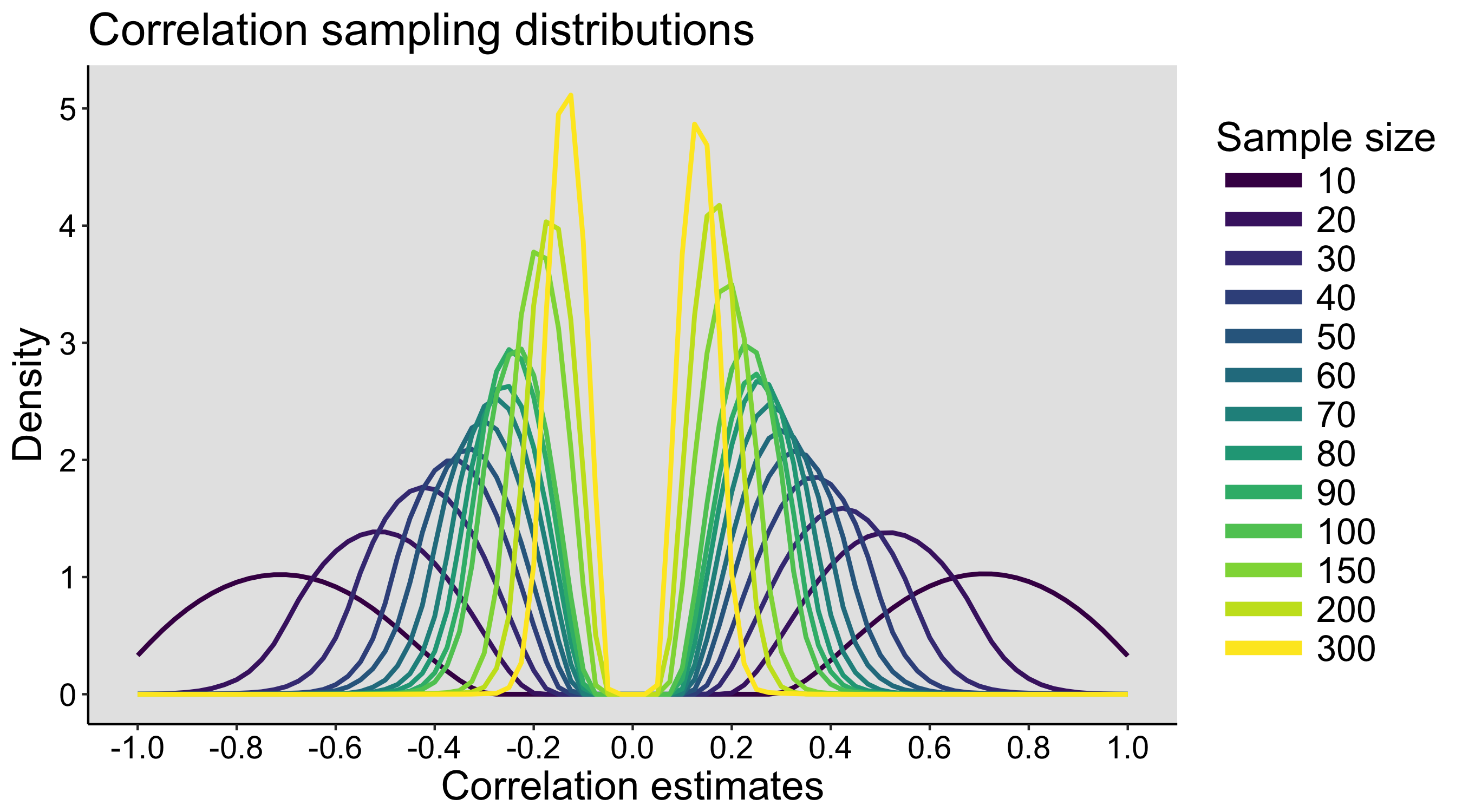
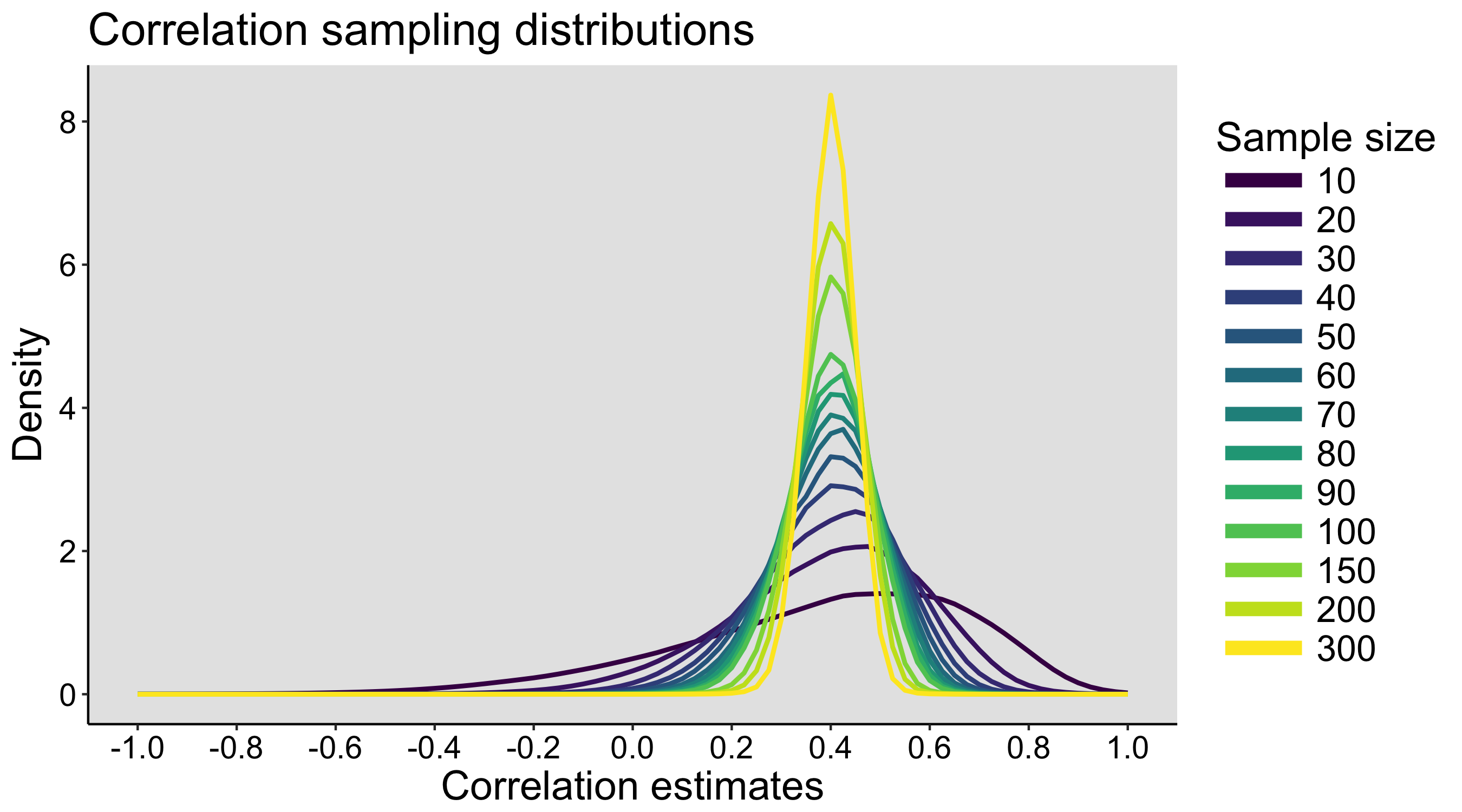
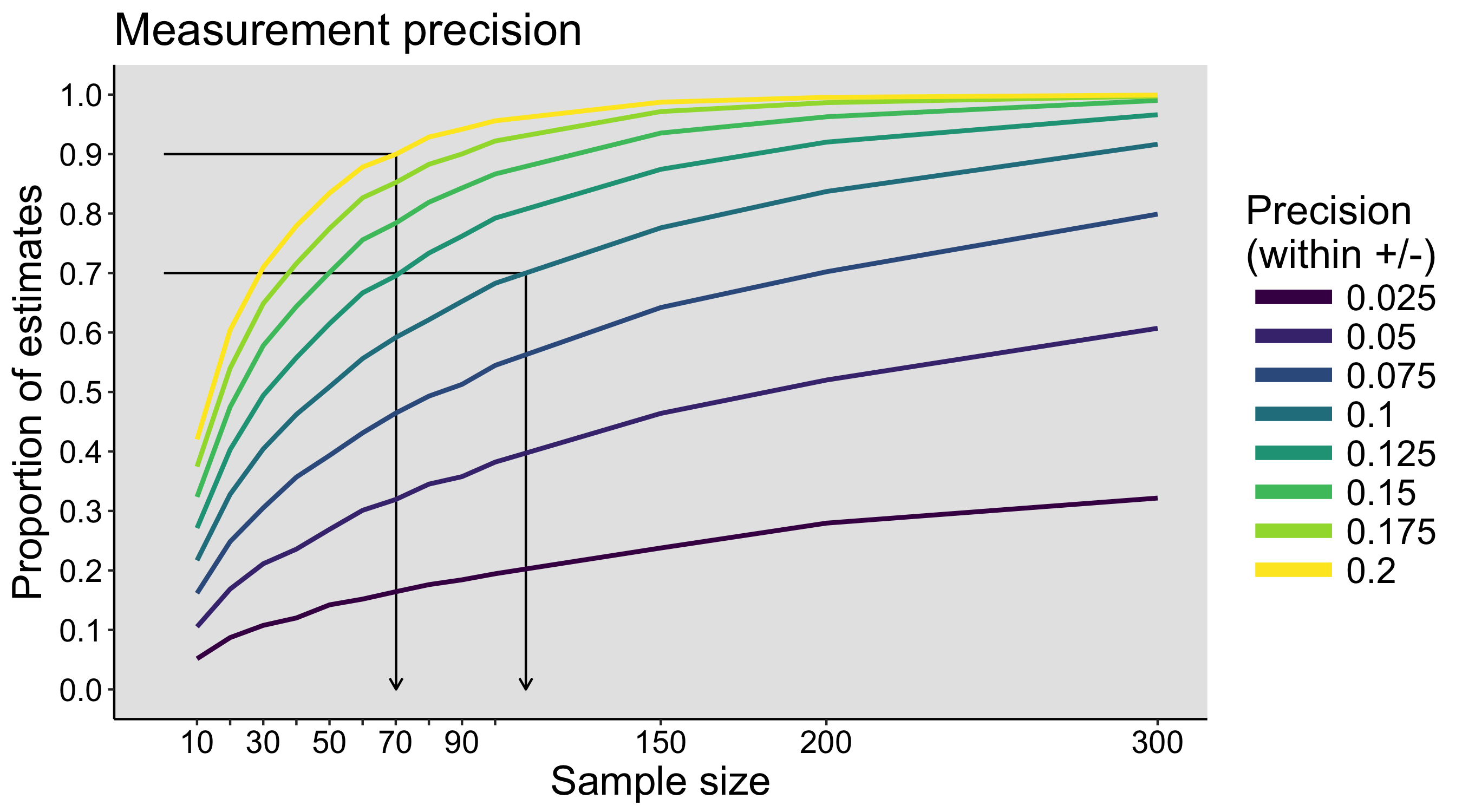
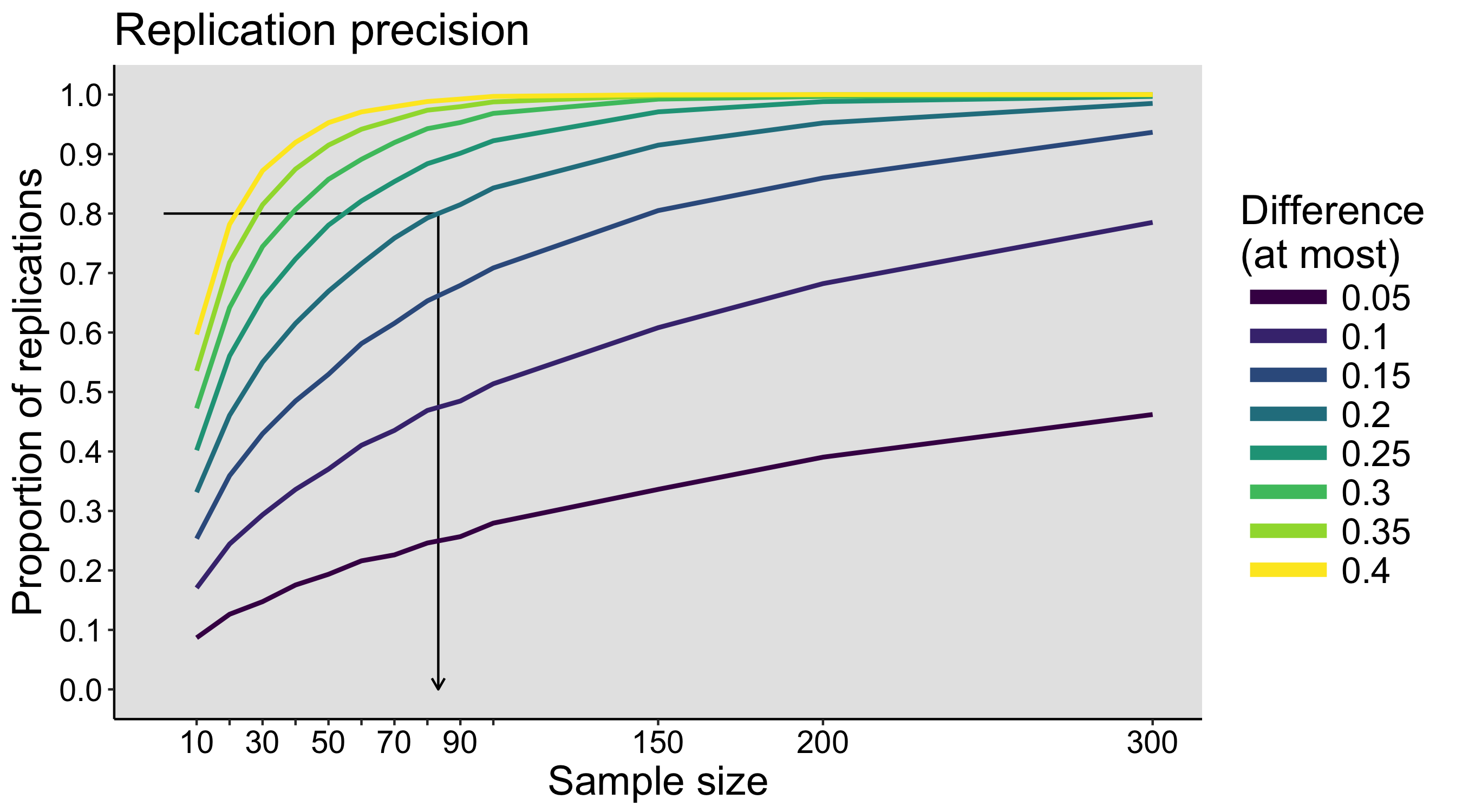
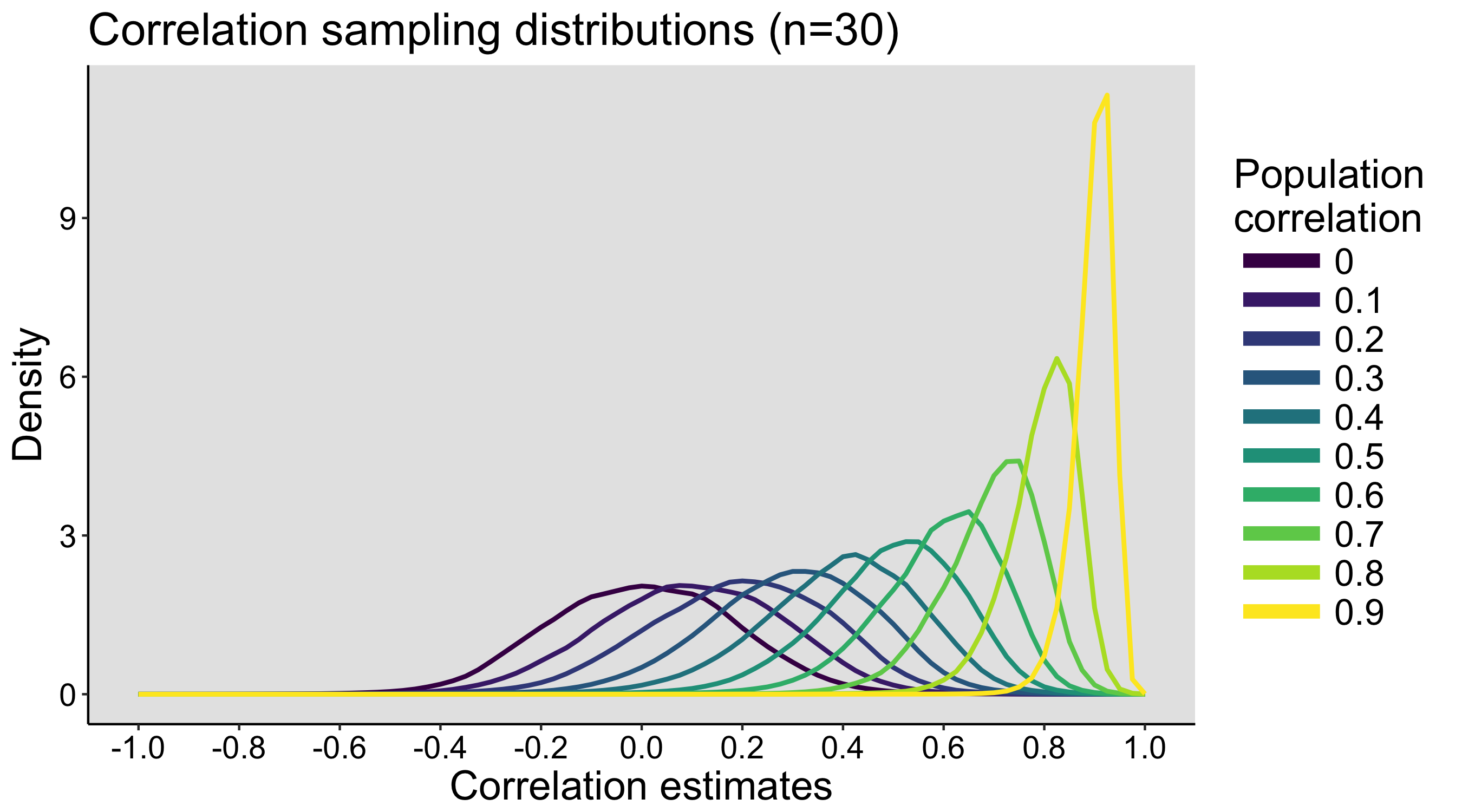
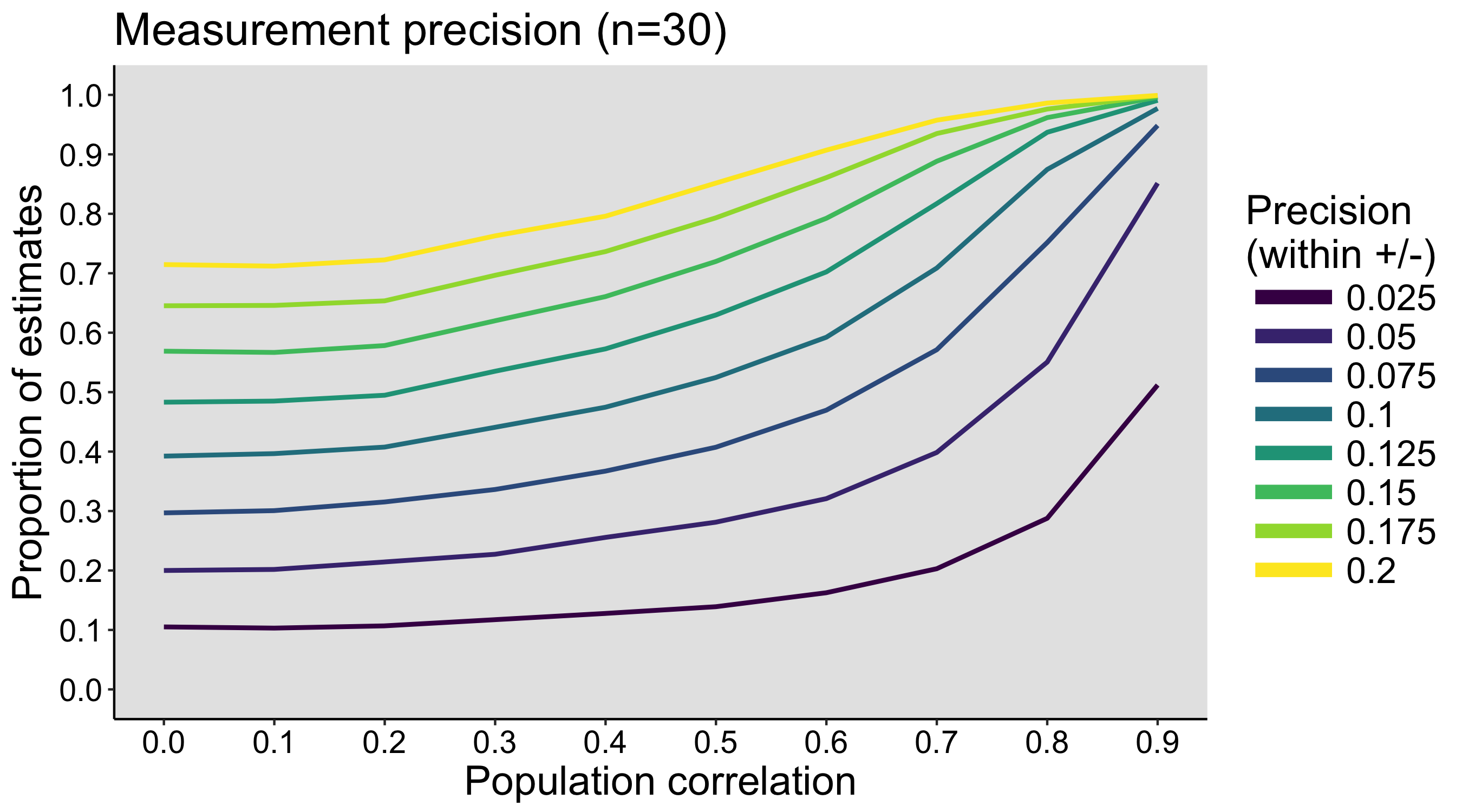
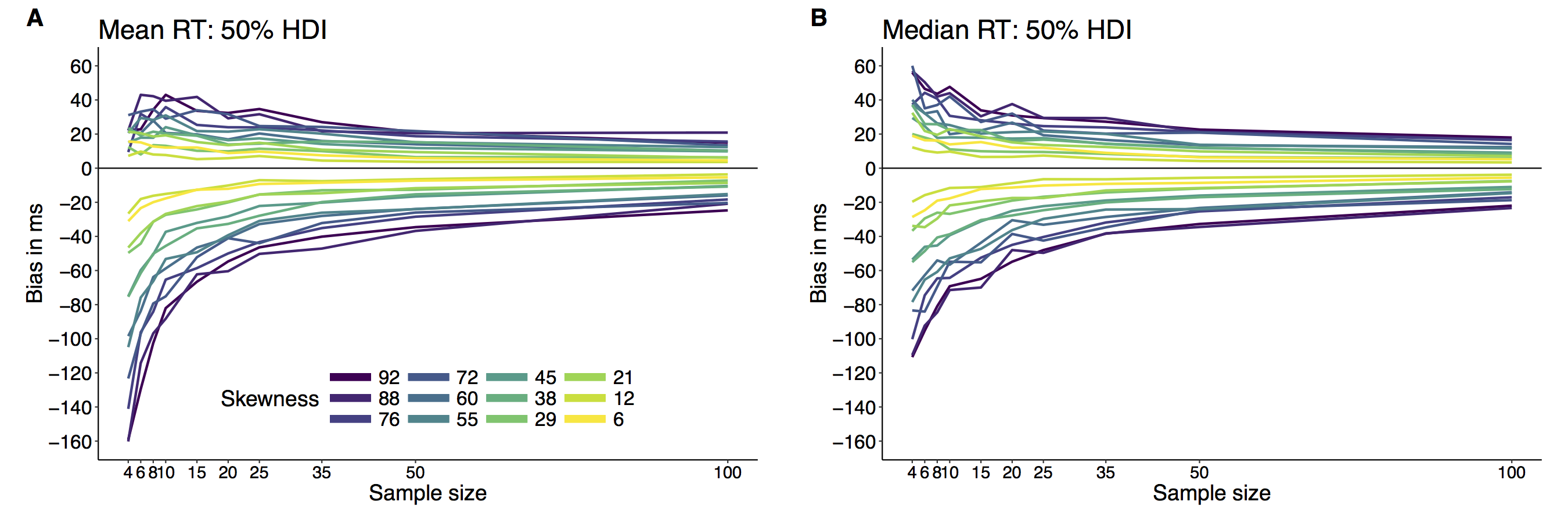
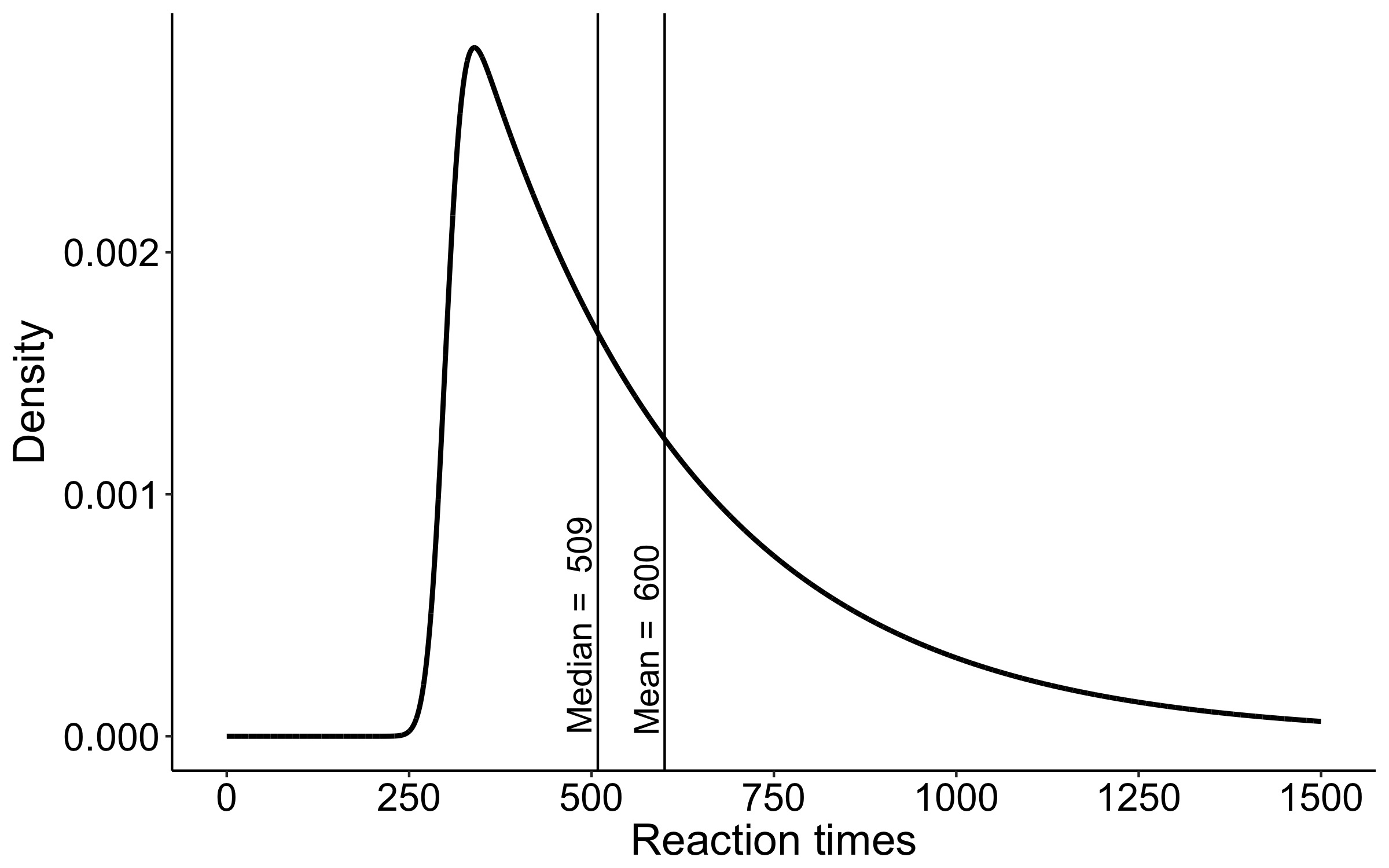
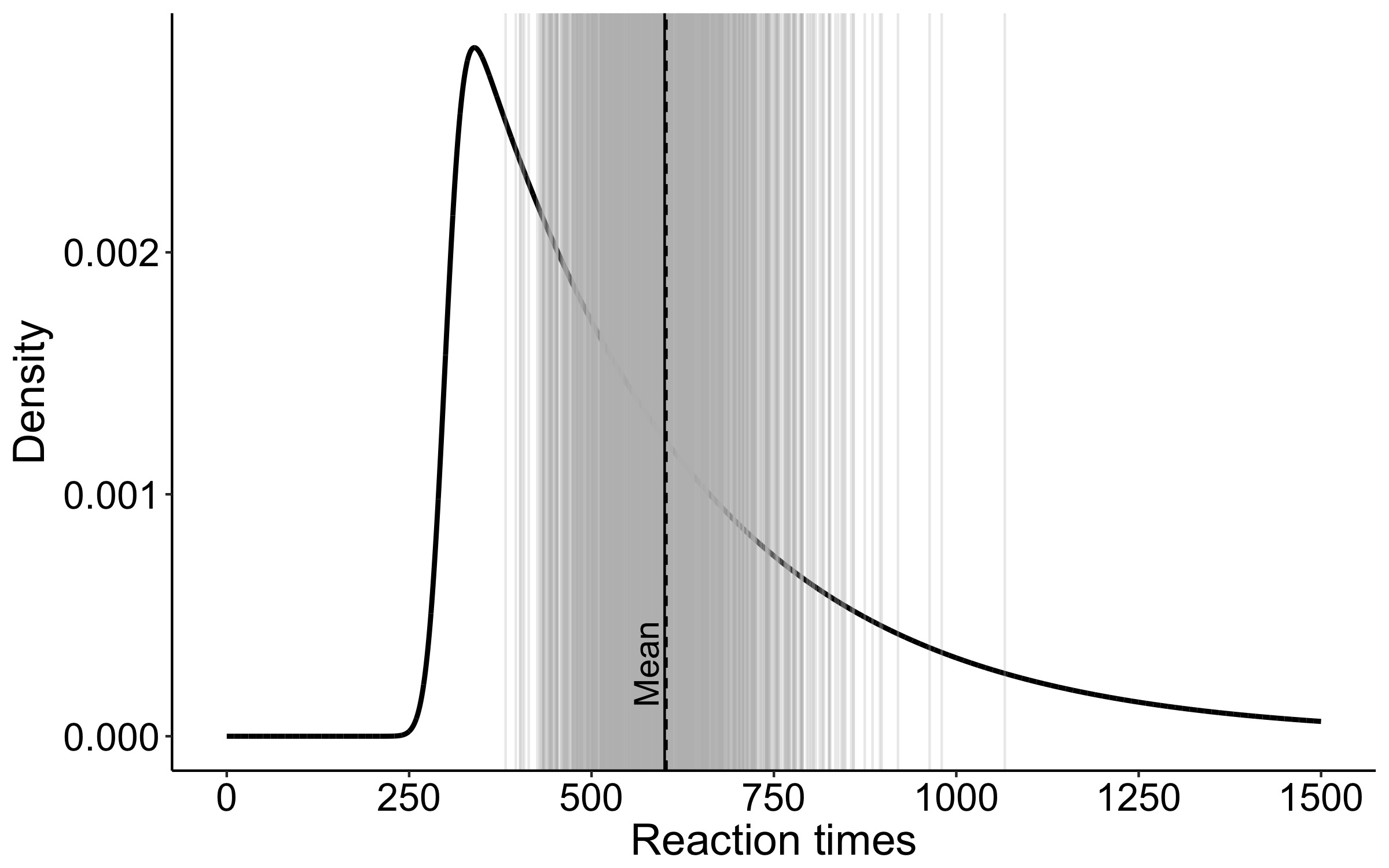
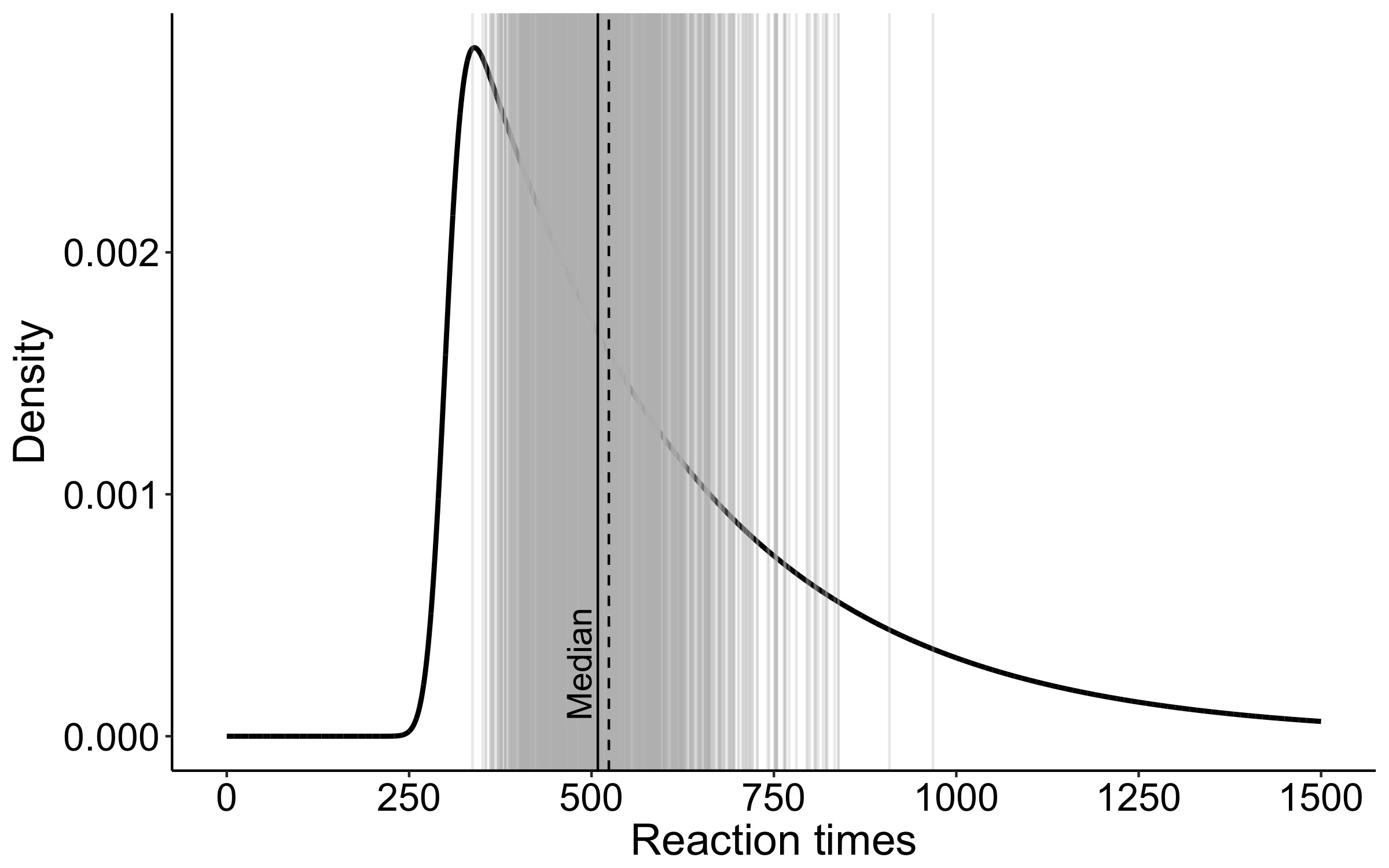
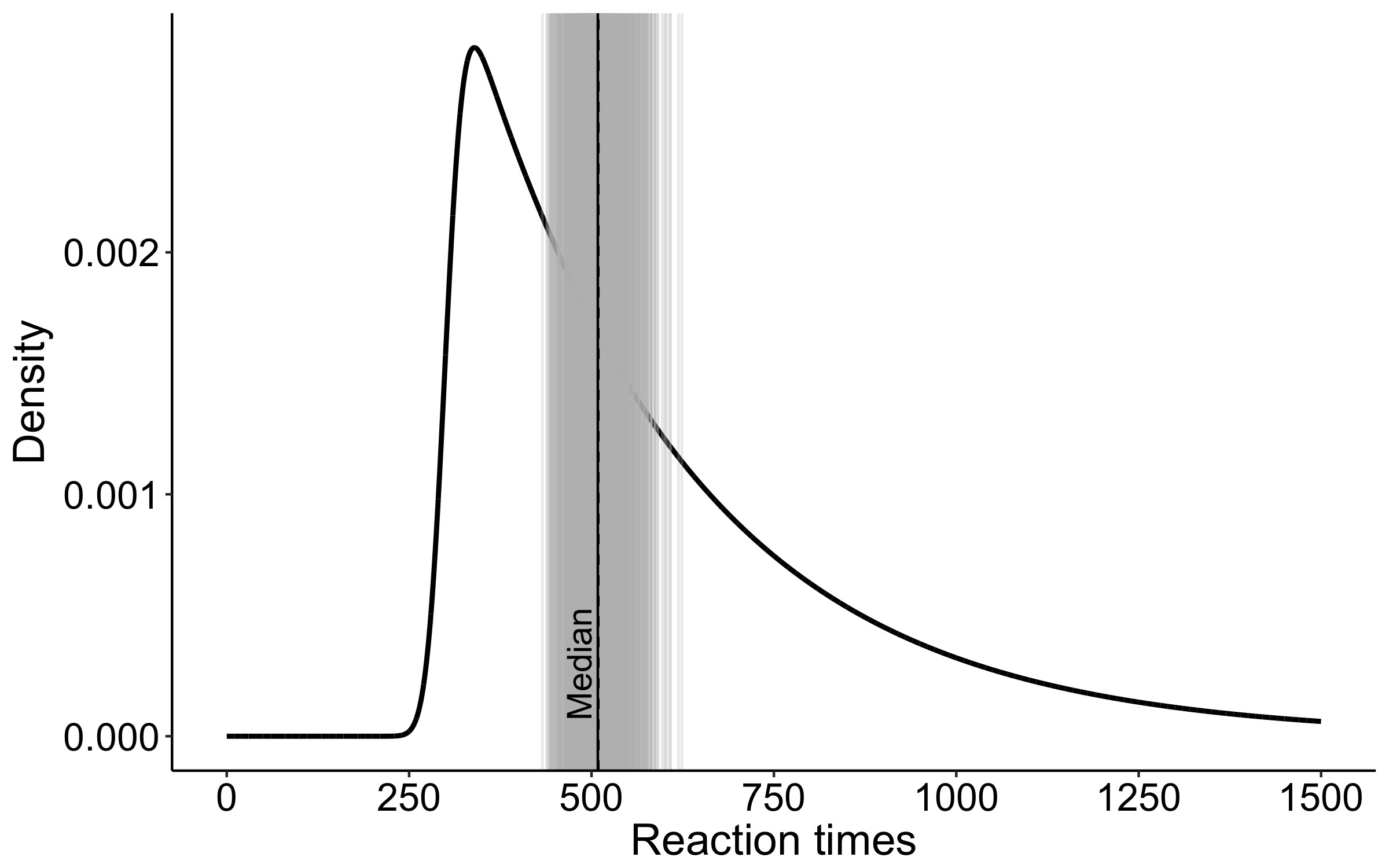
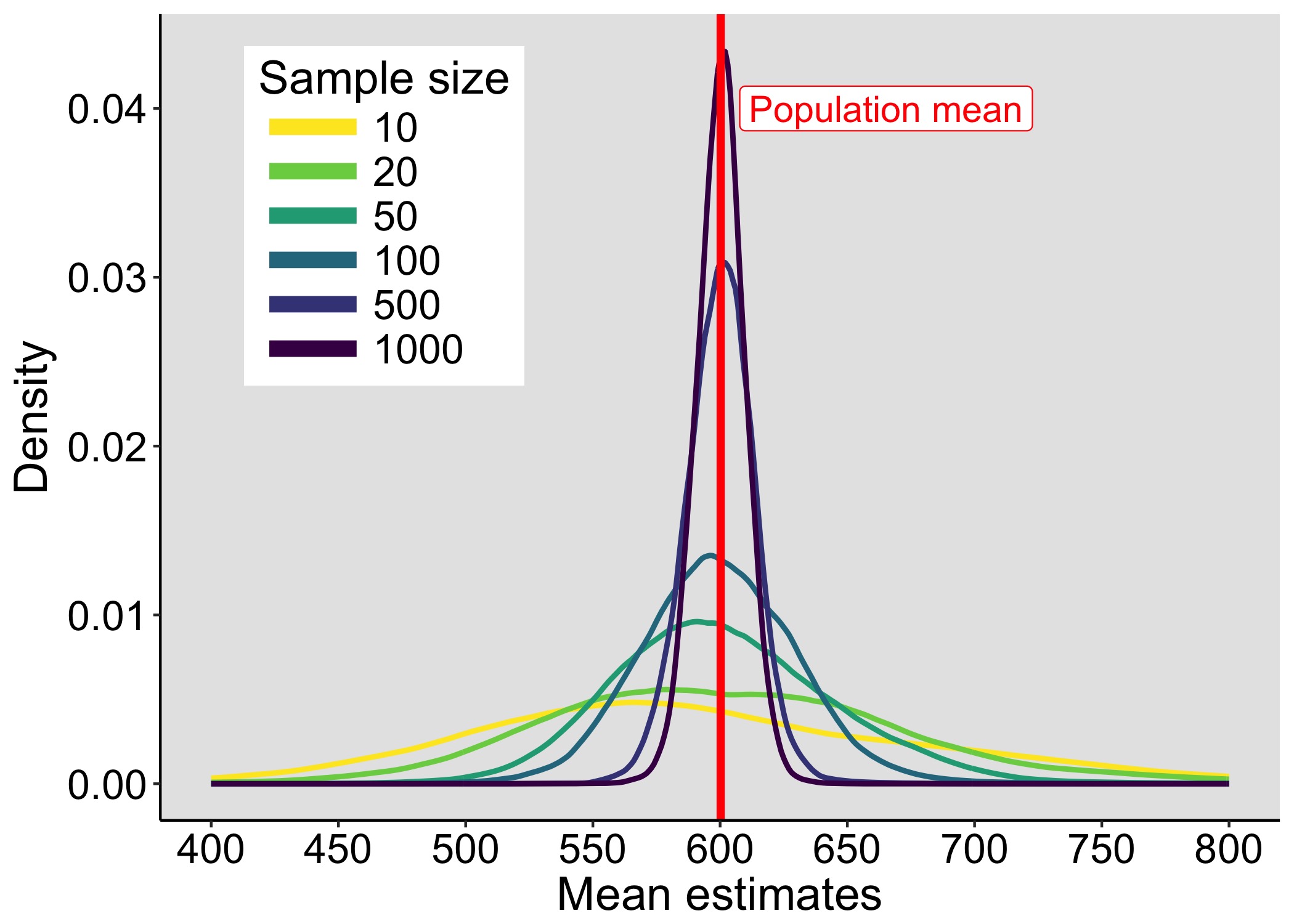
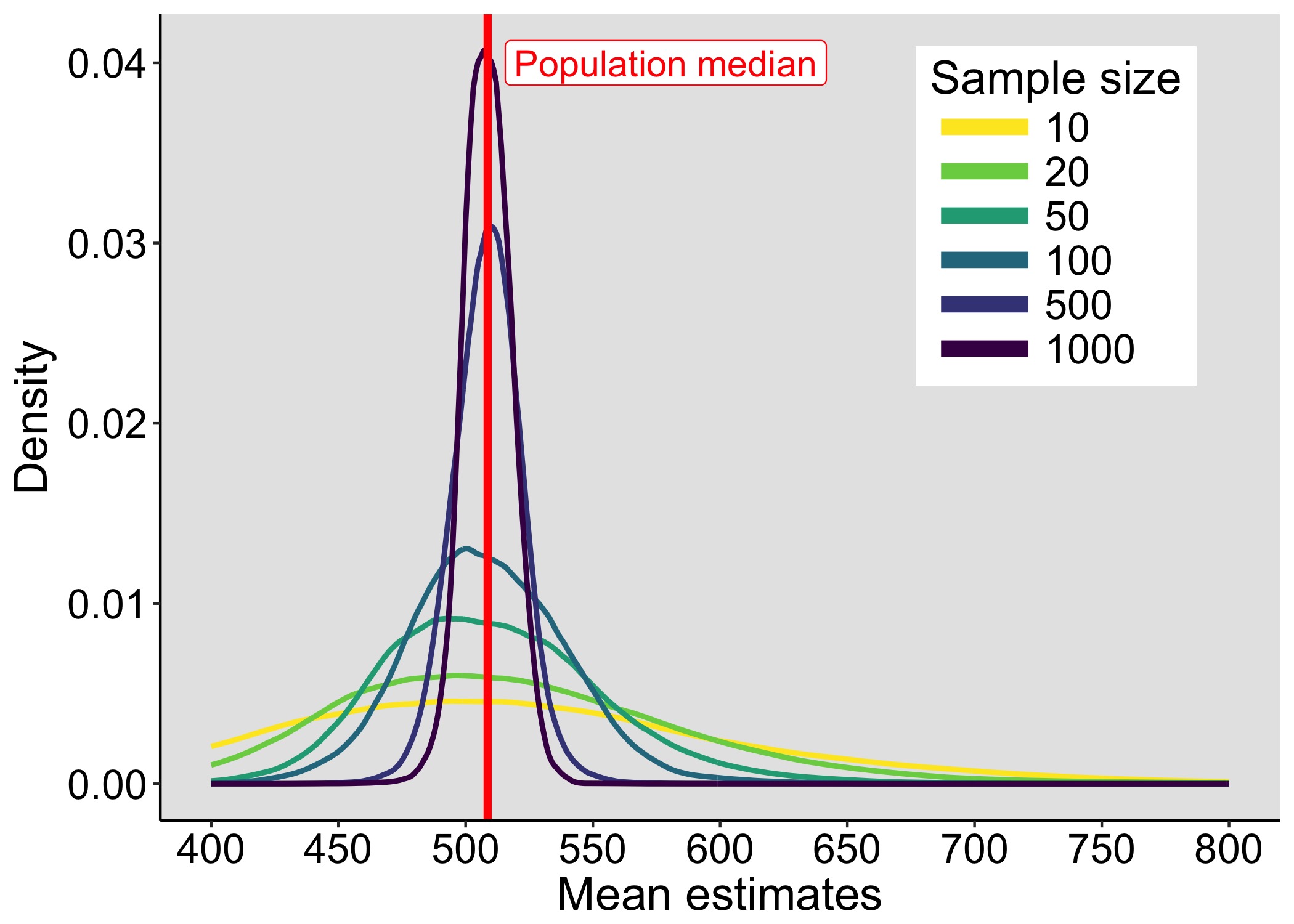
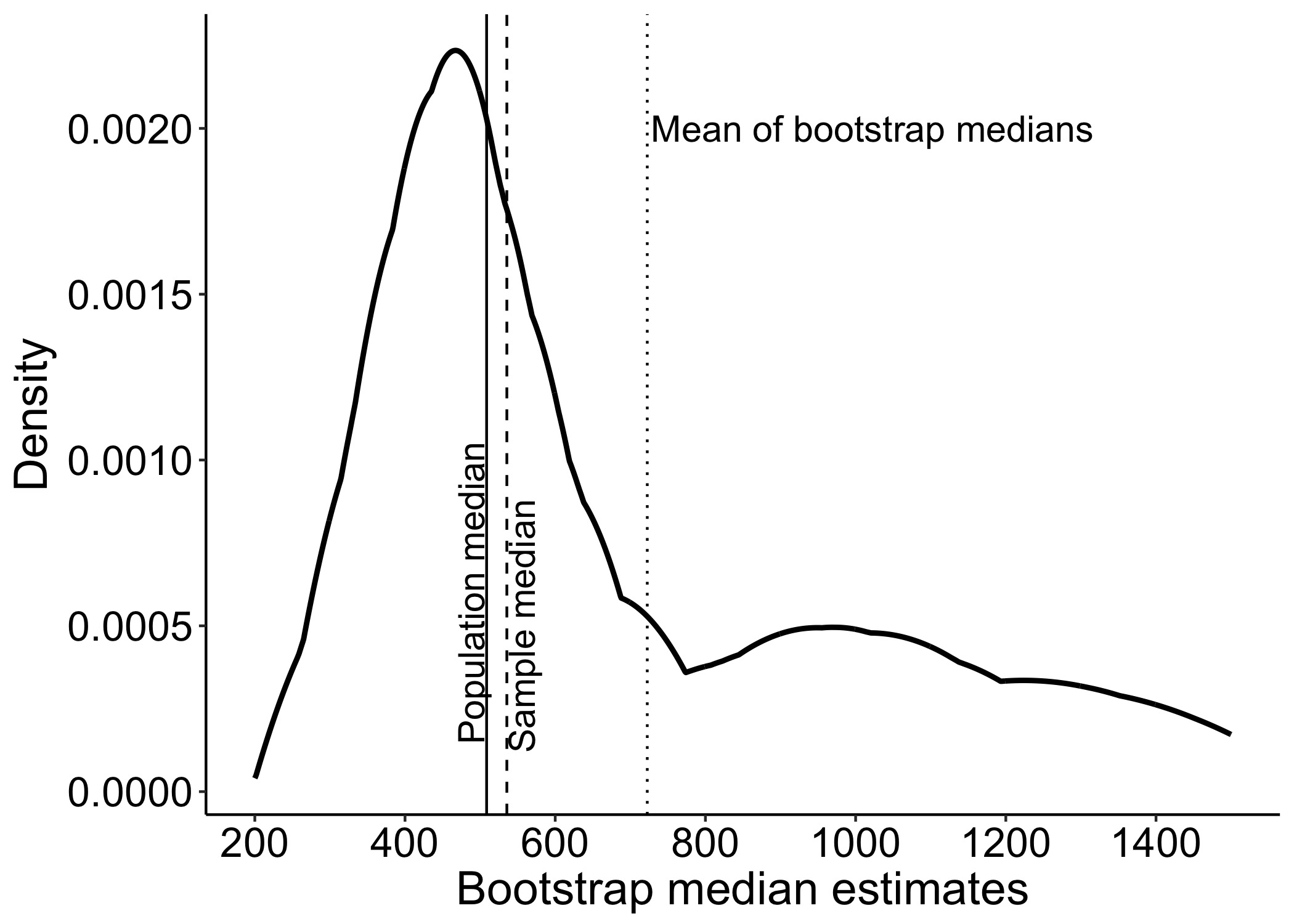
This year I’m teaching a new undergraduate course on the bootstrap for 4th year psychology students. Class examples, take-home exercises and the exam use R. I will also use a few analog activities in class. Here I’d like to share some of these activities. (This is also the opportunity to start a new category of posts on teaching.) The course is short, with only 5 sessions of 2 hours, but I think it is important to spend some of that time to try to get key concepts across by providing engaging activities. I’ll report back on how it went.
The 3 main activities involve dice, poker chips and wooden fish, to explore different types of sampling, sampling distributions, the distinction between sample and population, resampling…
Activity 1: dice (hierarchical sampling)
We use dice to simulate sampling with replacement from an infinite population of participants and trials.
This exercise provides an opportunity to learn about:
- the distinction between population and sample;
- sampling with replacement;
- hierarchical sampling;
- running simulations;
- estimation;
- the distinction between finite and infinite populations.
Material:
- 3 bags of dice
- 3 trays (optional)

Each bag contains a selection of dice with 4 to 20 facets, forming 3 independent populations. I used a lot of dice in each of bag but that’s not necessary. It just makes it harder to guess the content of the bags. I got the dice from the TheDiceShopOnline.
Many exercises can be proposed, involving different sampling strategies, with the aim of making some sort of inference about the populations. Here is the setup we will use in class:
- 3 participants or groups of participants are involved, each working independently with their own bag/population;
- a dice is randomly picked from a bag (without looking inside the bag!) — this is similar to randomly sampling a participant from the population;
- the dice is rolled 5 times, and the results written down — this is similar to randomly sampling trials from the participant;
- perform the two previous steps 10 times, for a total of 10 participants x 5 trials = 50 trials.

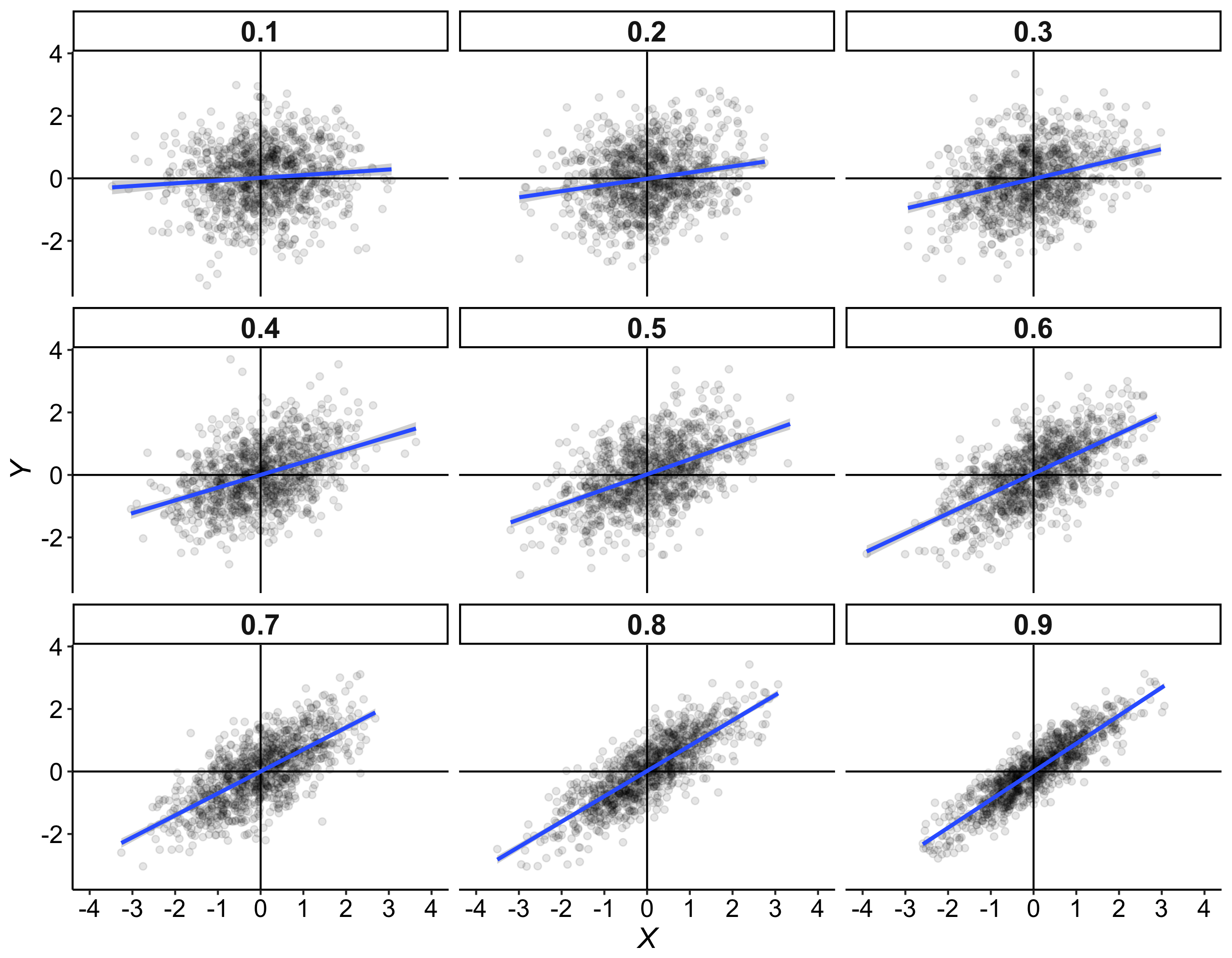
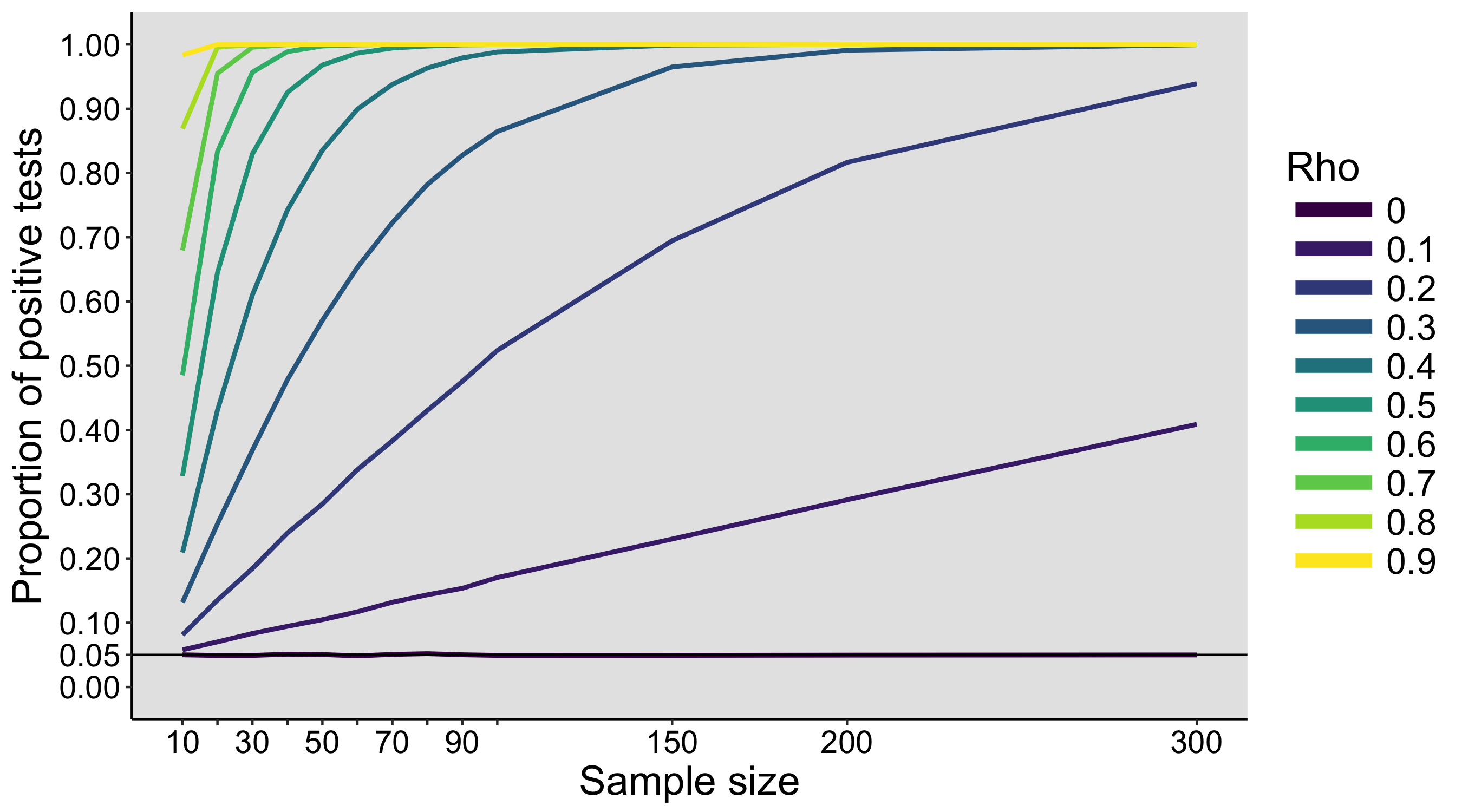
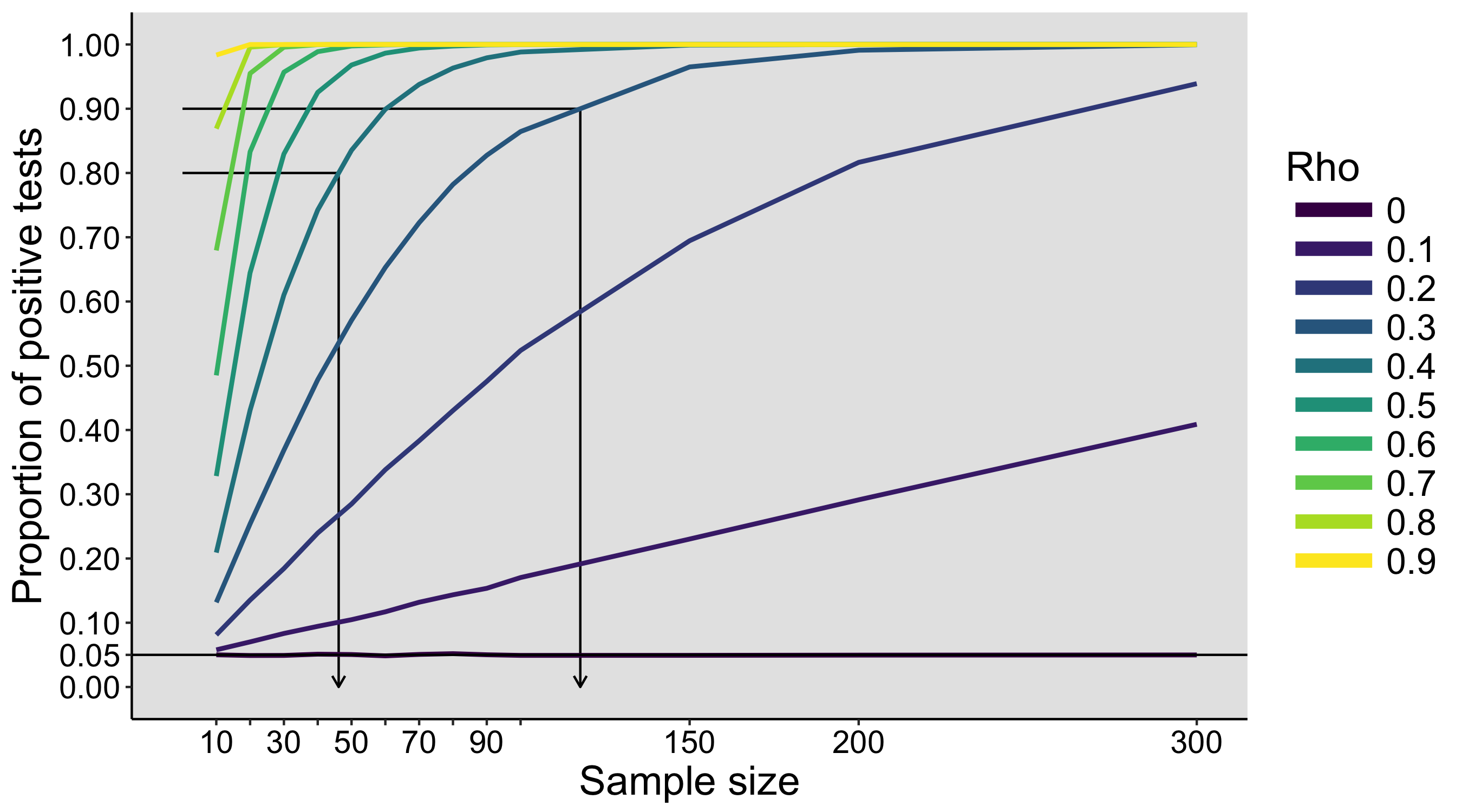
These values are then entered into a text file and shared with the rest of the class. The text files are opened in R, and the main question is: is there evidence that our 3 samples of 10 participants x 5 trials were drawn from different populations? To simplify the problem, a first step could involve averaging over trials, so we are left with 10 values from each group. The second step is to produce some graphical representation of the data. Then we can try various inferential statistics.
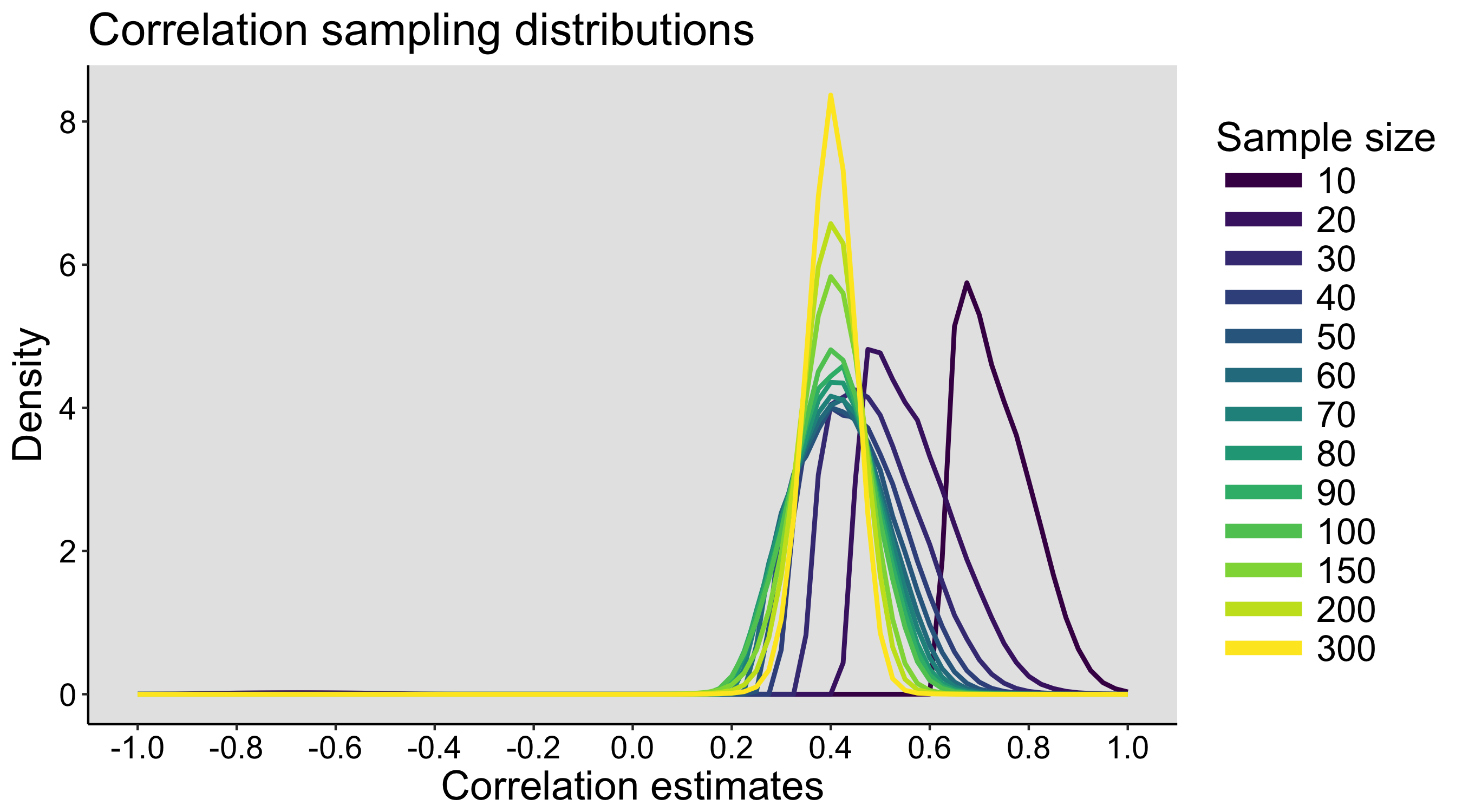
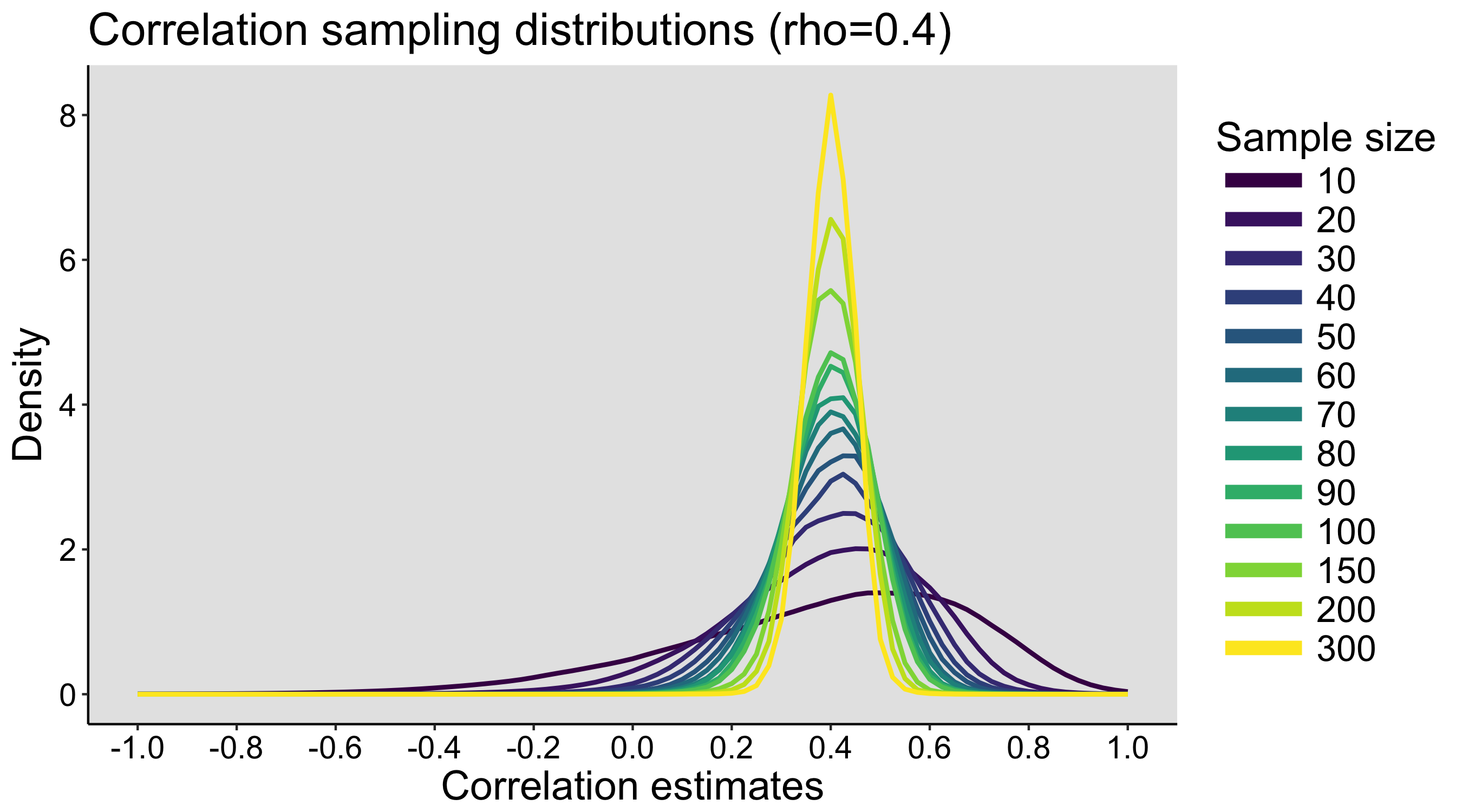
The exercise can be repeated on different days, to see how much variability we get between simulated experiments. During the last class, the populations and the sampling distributions are revealed.
Also, in this exercise, because the dice are sampled with replacement, the population has an infinite size. The content of each bag defines the probability of sampling each type of dice, but it is not the entire population, unlike in the faux fish activity (see below).
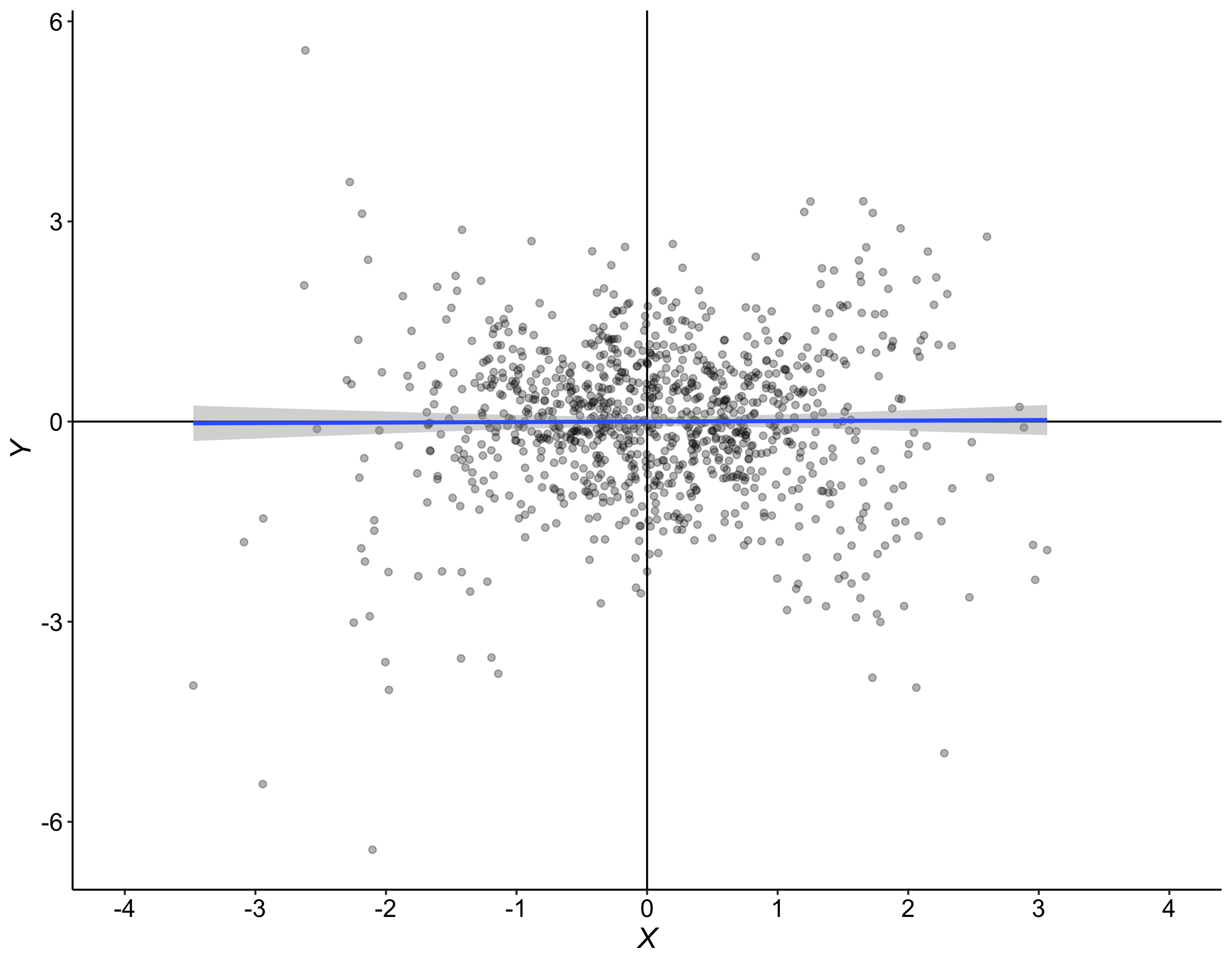
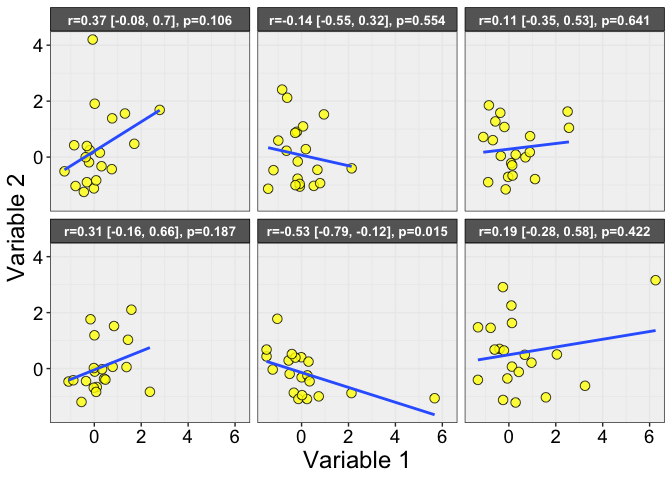
Here is an example of samples after averaging the 5 trials per dice/participant:

Activity 2: poker chips (bootstrap sampling with replacement)
We use poker chips to demonstrate sampling with replacement, as done in the bootstrap.
This exercise provides an opportunity to learn about:
- sampling with replacement;
- bootstrap sampling;
- running simulations.
A bag contains 8 poker chips, representing the outcome of an experiment. Each chip is like an observation.

First, we demonstrate sampling with replacement by getting a random chip from the bag, writing down its value, and replacing the chip in the bag. Second, we demonstrate bootstrap sampling by performing sampling with replacement 8 times, each time writing down the value of the random chip selected from the bag. This should help make bootstrap sampling intuitive.
After this analog exercise, we switch to R to demonstrate the implementation of sampling with replacement using the sample function.
Activity 3: faux fish (sampling distributions)
We sample with replacement from a finite population of faux fish to demonstrate the effect of sample size on the shape of sampling distributions.
The faux fish activity is mentioned in Steel, Liermann & Guttorp (2019), with pictures of class results. The activity is described in detail in Kelsey & Steel (2001).

This exercise provides an opportunity to learn about:
- the distinction between population and sample;
- sampling with replacement;
- running simulations;
- estimation;
- sampling distributions.
Material:
- two sets of 97 faux fish = fish-shaped bits of paper or other material
- two containers = ponds
- two large blank sheets of paper
- x axis = ‘Mean weight (g)’
- y axis = ‘Number of experiments’
- titles = ‘n=3 replicates’ / ‘n=10 replicates’
I got faux fish made of wood from Wood Craft Shapes.

Each faux fish has a weight in grams written on it.

The frequencies of the weights is given in Kelsey & Steel (2001).

The fish population is stored in a box. I made 2 identical populations, so that two groups can work in parallel.

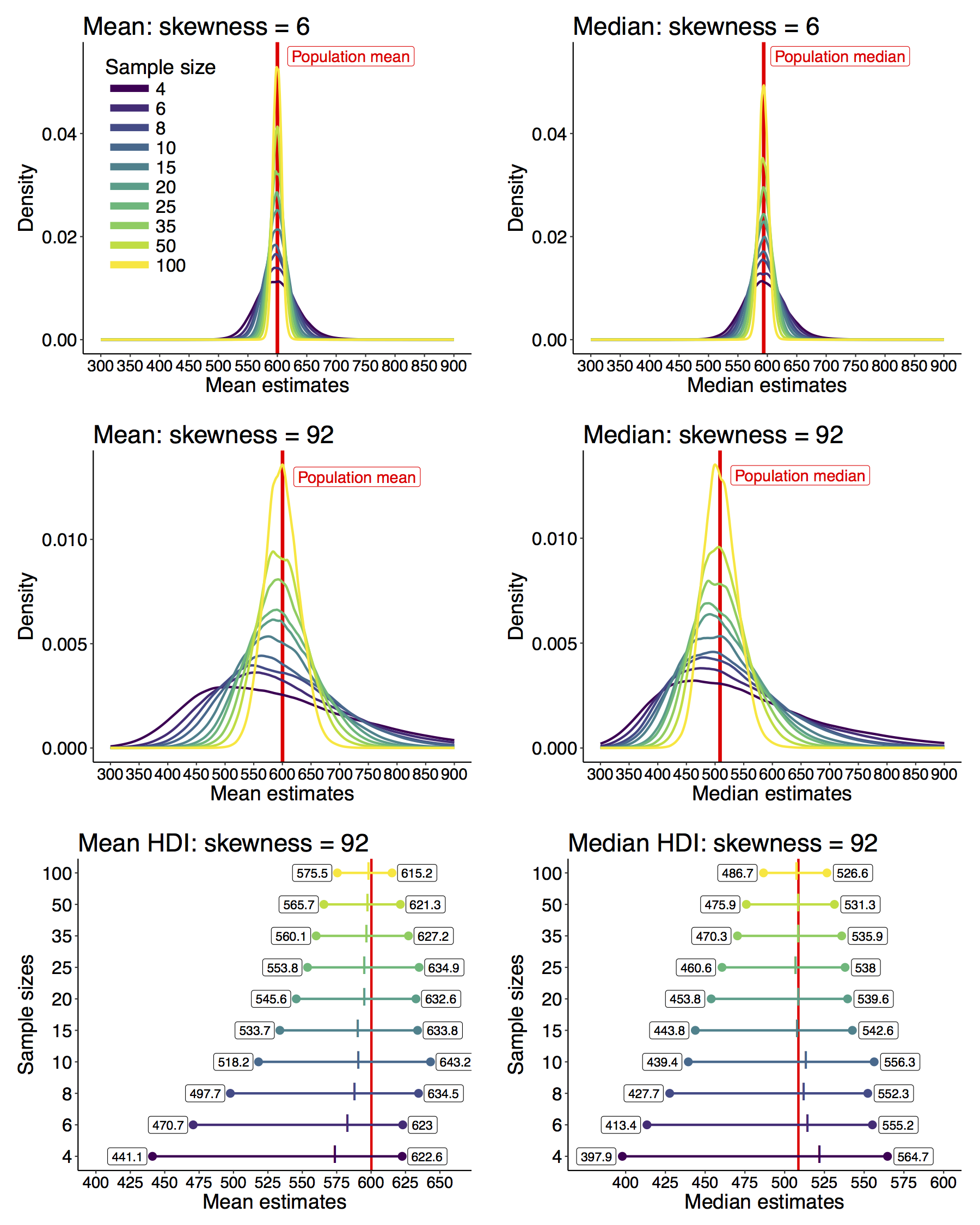
The first goal of the exercise is to produce sampling distributions by sampling with replacement from a population. The second goal is to evaluate the effect of the sample size on the shape of the sampling distribution. The third goal is to experiment with a digital version of the analog task, to gain familiarity with simulations.
Unlike the dice activity, this activity involves a finite size population: each box contains the full population under study.
Setup:
- two groups of participants;
- each group is assigned a box;
- participants from each group take turn sampling from the box n=3 or n=10 faux fish (depending on the group), without looking inside the box;
- each participant averages the numbers, writes down the answer and marks it on the large sheet of paper assigned to each group;
- this is repeated until a sufficient number of simulated experiments have been carried out to assess the shape of the resulting sampling distribution.
To speed up the exercise, a participant picks n fish, writes down the weights, puts all the fish back in the box, and passes the box to the next participant. While the next participant is sampling fish from the box, the previous participant computes the mean and marks the result on the group graph.
Once done, the class discusses the results:
- the sampling distributions are compared;
- the population mean is revealed;
- the population is revealed by showing the handout from the book and opening the boxes.
Then we do the same in R, but much quicker!
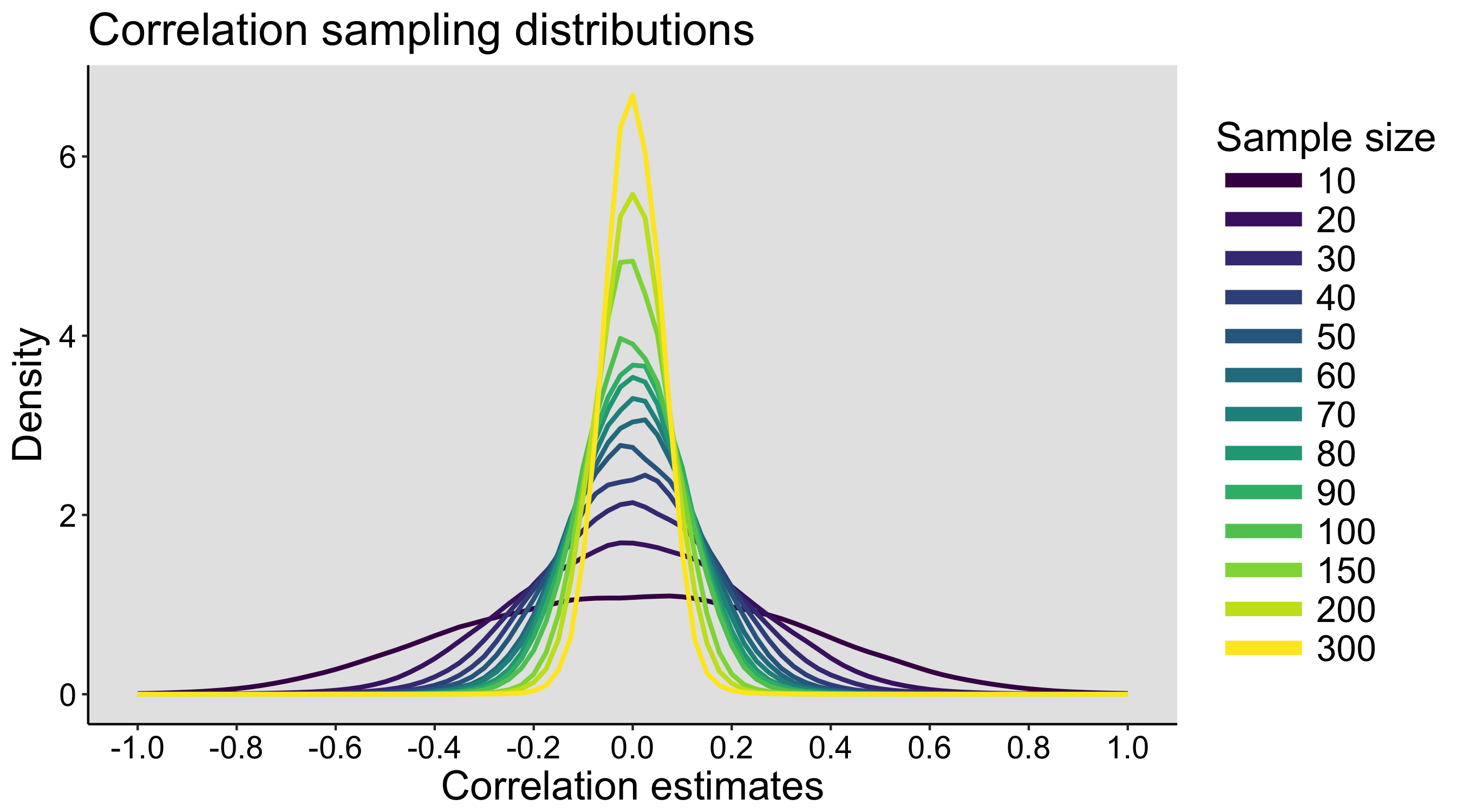
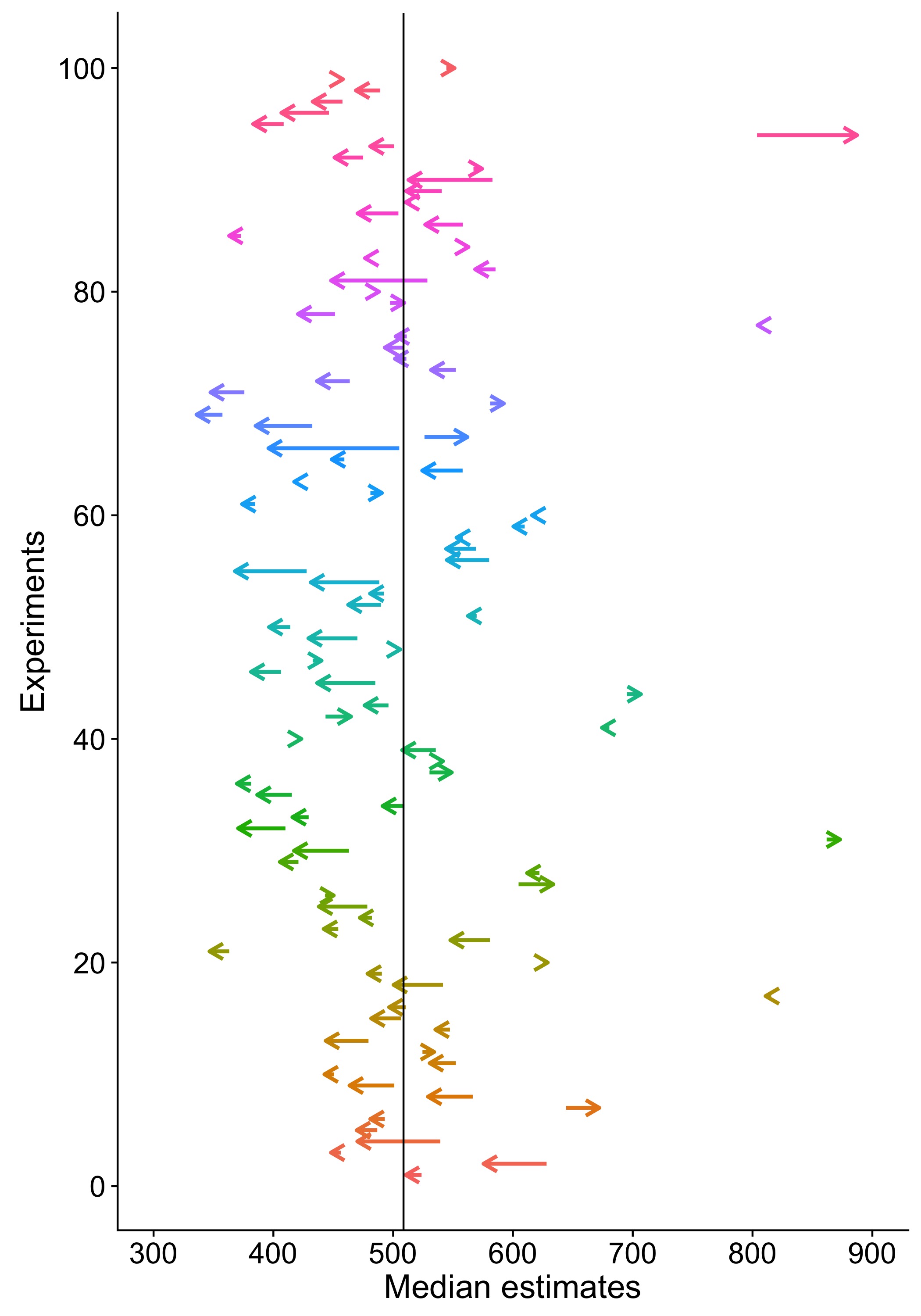
Here is an example of simulated results for n=3 (the vertical line marks the population mean):

References
Kelsey, Kathryn, and Ashley Steel. The Truth about Science: A Curriculum for Developing Young Scientists. NSTA Press, 2001.
Steel, E. Ashley, Martin Liermann, and Peter Guttorp. Beyond Calculations: A Course in Statistical Thinking. The American Statistician 73, no. sup1 (29 March 2019): 392–401. https://doi.org/10.1080/00031305.2018.1505657.