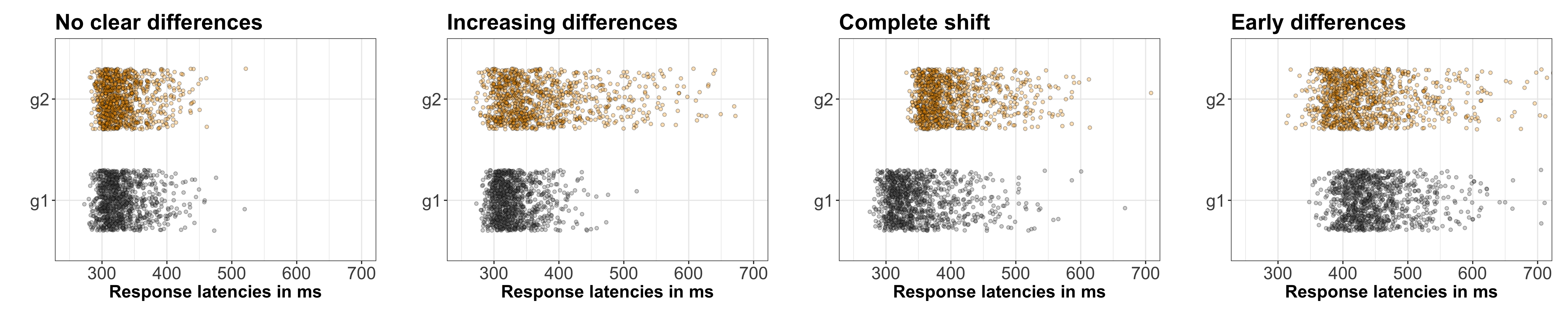
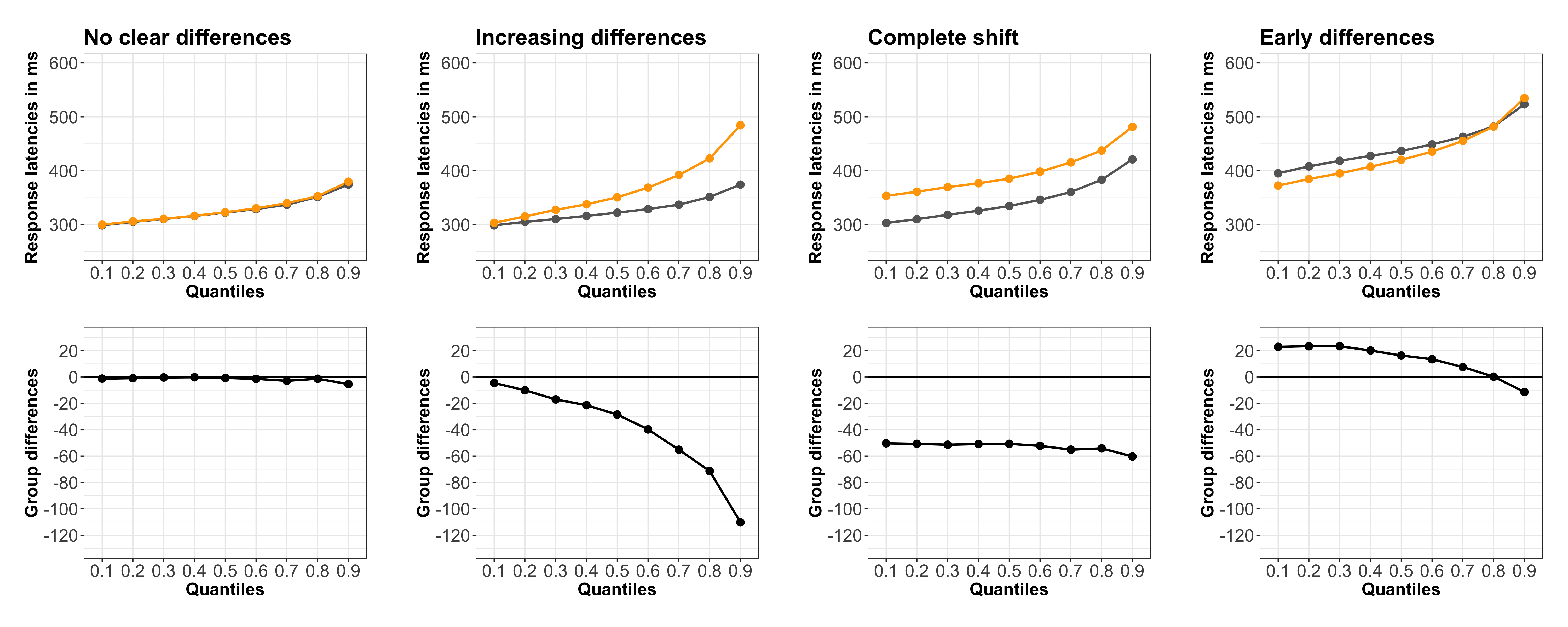
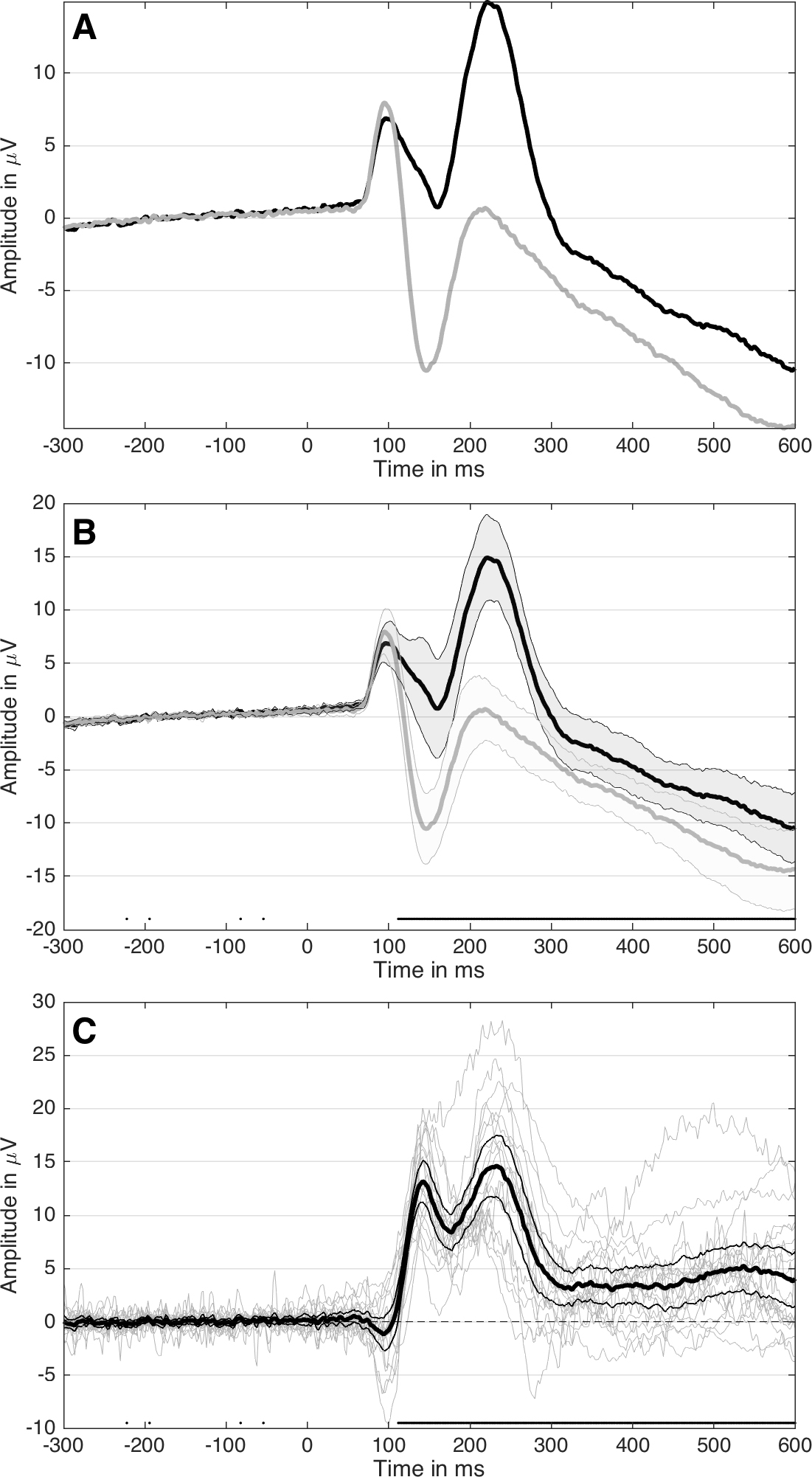
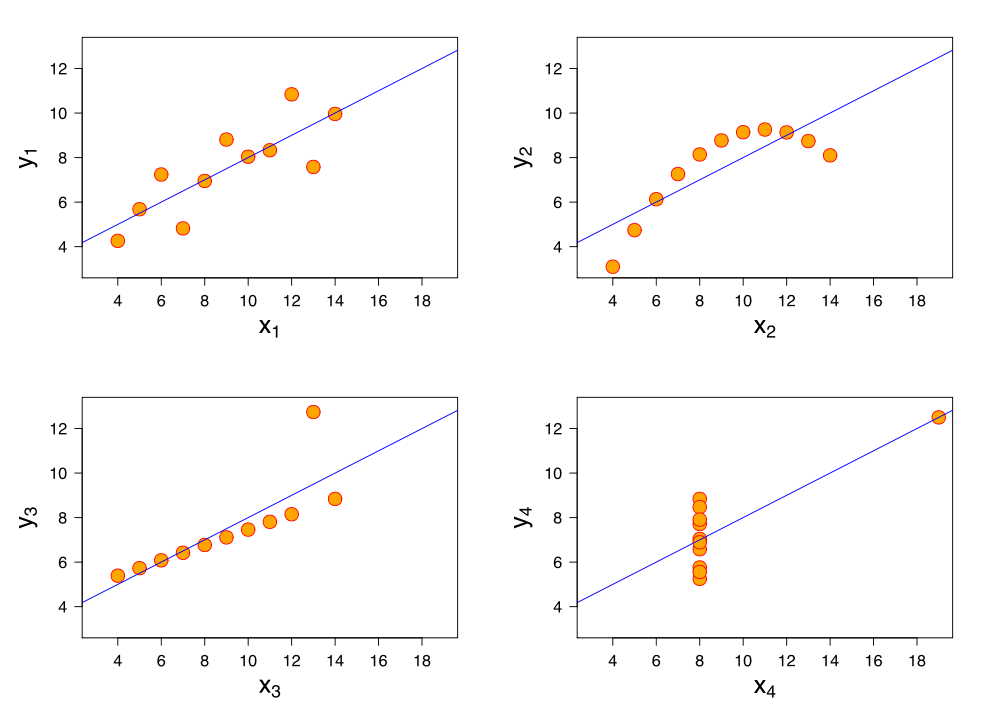
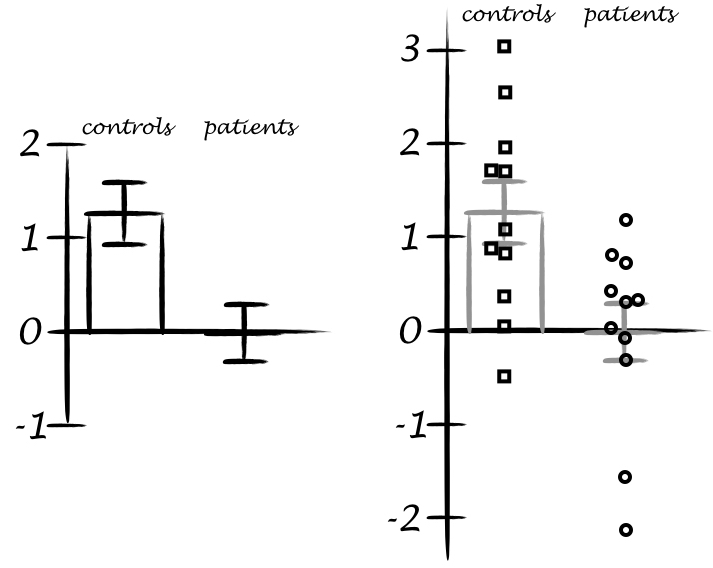
Many methods have been proposed to assess the success of a replication (Costigan et al. 2024; Cumming & Maillardet 2006; Errington et al. 2021; LeBel et al. 2019; Ly et al. 2019; Mathur & VanderWeele 2020; Muradchanian et al., 2021; Patil et al. 2016; Spence & Stanley 2024; Verhagen & Wagenmakers 2014). The most common method, also used to determine if results from similar experimental designs are consistent across studies, is consistency in statistical significance: do the two studies report a p value less than some (usually) arbitrary threshold? This approach can be misleading for many reasons, for instance when two studies report the same group difference, but different confidence intervals: one including the null, the other one excluding it. Even though the group differences are the same, sampling error combined with statistical significance would lead us to conclude that the two studies disagree. There is a very nice illustration of the issue in Figure 1 of Amrhein, Greenland & McShane (2019).
More generally:
“if the alternative is correct and the actual power of two studies is 80%, the chance that the studies will both show P ≤ 0.05 will at best be only 0.80(0.80) = 64%; furthermore, the chance that one study shows P ≤ 0.05 and the other does not (and thus will be misinterpreted as showing conflicting results) is 2(0.80)0.20 = 32% or about 1 chance in 3.”
Greenland et al. 2016 (see also Amrhein, Trafimow & Greenland 2019)
So, in the long run, even if two studies always sample from the same population (even assuming all unmeasured sources of variability are the same across labs; Gelman et al. 2023), the literature would look like there is a replication crisis when none exists.
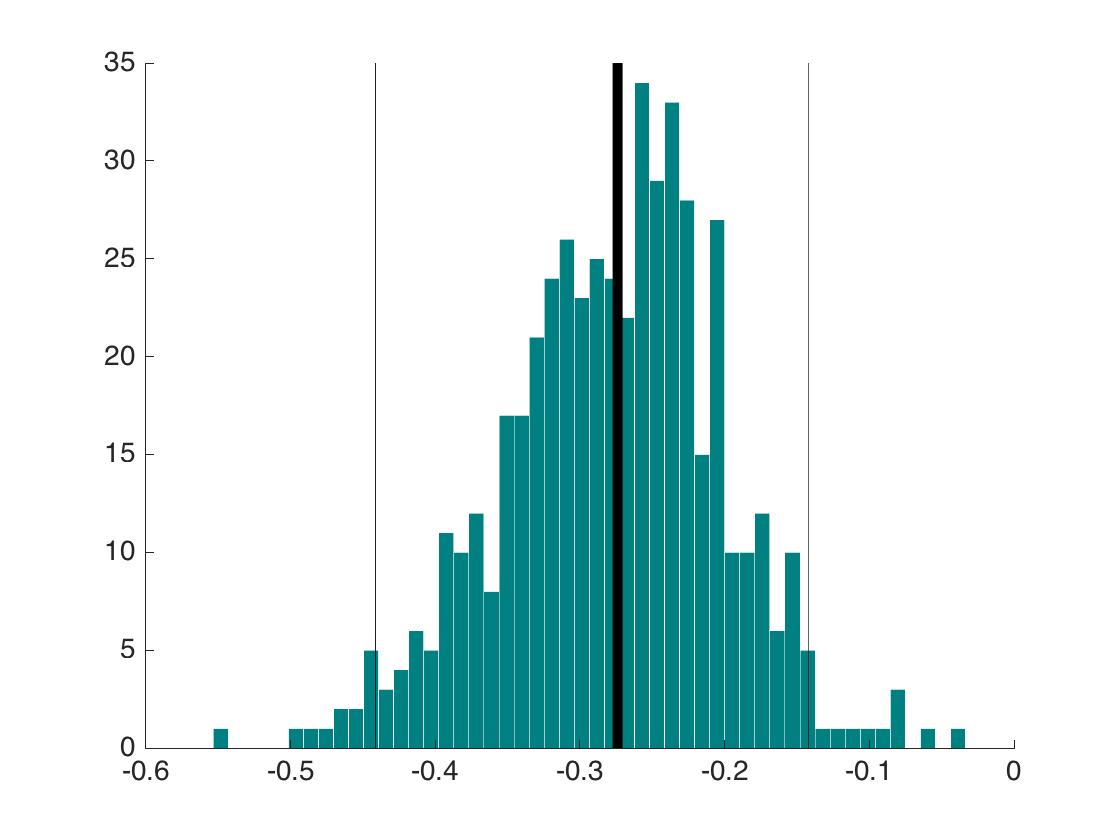
Let’s expand the single values from the example by Greenland et al. (2016) and plot the probability of finding consistent and inconsistent results as a function of power:

When deciding about consistency between experiments using the statistical significance criterion, the probability to reach the correct decision depends on power, and unless power is very high, we will often be wrong.
In the previous figure, why consider power as low as 5%? If that seems unrealistic, a search for n=3 or n=4 in Nature and Science magazines will reveal recent experiments carried out with very small sample sizes in the biological sciences. Also, in psychology, interactions require much larger sample sizes than typically used, for instance when comparing correlation coefficients (Rousselet, Pernet & Wilcox, 2023). So very low power is still a real concern.
In practice, the situation is probably worse, because power analyses are typically performed assuming parametric assumptions are met; so the real power of a line of research will be lower than expected — see simulations in Rousselet & Wilcox (2020); Wilcox & Rousselet (2023); Rousselet, Pernet & Wilcox (2023).
To provide an illustration of the effect of skewness on power, and in turn, on replication success based on statistical significance, let’s use g-and-h distributions — see details in Rousselet & Wilcox (2020) and Yan & Genton (2019). Here we consider h=0 and vary g from 0 (normal distribution) to 1 (shifted lognormal distribution):

Now, let’s do a simulation in which we vary g, take samples of n=20, and perform a one-sample t-test on means, 10% trimmed means and 20% trimmed means. The code is on GitHub. To assess power, a constant is added to each sample, assuming a power of 80% when sampling from a standard normal population (g=h=0). Alpha is set to the arbitrary value of 0.05. The simulation includes 100,000 iterations.
Here are the results for false positives, showing a non-linear increase as a function g, with the one-sample t-test much more affected when using means than trimmed means:

And the true positive results, showing lower power for trimmed means under normality, but much more resilience to increasing skewness than the mean.

These results are well known (see for instance Rousselet & Wilcox, 2020).
Now the novelty is to consider in turn the impact on the probability of a positive outcome in both experiments.

If we assume normality and determine our sample size to achieve 80% power in the long run, skewness can considerably lower the probability of observing two studies both showing p<0.05 if we employ a one-sample t-test on means. Trimmed means are much less affected by skewness. Other robust methods will perform even better (Wilcox & Rousselet, 2023).
In the same setting, here is the probability of a positive outcome in one experiment and a negative outcome in the other one:

Let’s consider h = 0.1, so that outliers are more likely than in the previous simulation:

In the presence of outliers, false positives increase even more with g for the mean:

And power is overall reduced for all methods:

This reduction in power leads to even lower probability of consistent results than in the previous simulation:

And here are the results on the probability of observing inconsistent results:

So in the presence of skewness and outliers, the situation is overall even worse than suggested by Greenland et al. (2016). For this and other reasons, consistency in statistical significance should not be used to infer the success of a replication.
References
Amrhein, V., Greenland, S., & McShane, B. (2019). Scientists rise up against statistical significance. Nature, 567(7748), 305. https://doi.org/10.1038/d41586-019-00857-9
Amrhein, V., Trafimow, D., & Greenland, S. (2019). Inferential Statistics as Descriptive Statistics: There Is No Replication Crisis if We Don’t Expect Replication. The American Statistician, 73(sup1), 262–270. https://doi.org/10.1080/00031305.2018.1543137
Costigan, S., Ruscio, J., & Crawford, J. T. (2024). Performing Small-Telescopes Analysis by Resampling: Empirically Constructing Confidence Intervals and Estimating Statistical Power for Measures of Effect Size. Advances in Methods and Practices in Psychological Science, 7(1), 25152459241227865. https://doi.org/10.1177/25152459241227865
Cumming, G., & Maillardet, R. (2006). Confidence intervals and replication: Where will the next mean fall? Psychological Methods, 11(3), 217–227. https://doi.org/10.1037/1082-989X.11.3.217
Errington, T. M., Mathur, M., Soderberg, C. K., Denis, A., Perfito, N., Iorns, E., & Nosek, B. A. (2021). Investigating the replicability of preclinical cancer biology. eLife, 10, e71601. https://doi.org/10.7554/eLife.71601
Gelman, A., Hullman, J., & Kennedy, L. (2023). Causal Quartets: Different Ways to Attain the Same Average Treatment Effect. The American Statistician. https://www.tandfonline.com/doi/full/10.1080/00031305.2023.2267597
Greenland, S., Senn, S. J., Rothman, K. J., Carlin, J. B., Poole, C., Goodman, S. N., & Altman, D. G. (2016). Statistical tests, P values, confidence intervals, and power: A guide to misinterpretations. European Journal of Epidemiology, 31(4), 337–350. https://doi.org/10.1007/s10654-016-0149-3
LeBel, E. P., Vanpaemel, W., Cheung, I., & Campbell, L. (2019). A Brief Guide to Evaluate Replications. Meta-Psychology, 3. https://doi.org/10.15626/MP.2018.843
Ly, A., Etz, A., Marsman, M., & Wagenmakers, E.-J. (2019). Replication Bayes factors from evidence updating. Behavior Research Methods, 51(6), 2498–2508. https://doi.org/10.3758/s13428-018-1092-x
Mathur, M. B., & VanderWeele, T. J. (2020). New Statistical Metrics for Multisite Replication Projects. Journal of the Royal Statistical Society Series A: Statistics in Society, 183(3), 1145–1166. https://doi.org/10.1111/rssa.12572
Muradchanian, J., Hoekstra, R., Kiers, H., & van Ravenzwaaij, D. (2021). How best to quantify replication success? A simulation study on the comparison of replication success metrics. Royal Society Open Science, 8(5), 201697. https://doi.org/10.1098/rsos.201697
Patil, P., Peng, R. D., & Leek, J. T. (2016). What should we expect when we replicate? A statistical view of replicability in psychological science. Perspectives on Psychological Science : A Journal of the Association for Psychological Science, 11(4), 539–544. https://doi.org/10.1177/1745691616646366
Rousselet, G., Pernet, C. R., & Wilcox, R. R. (2023). An introduction to the bootstrap: A versatile method to make inferences by using data-driven simulations. Meta-Psychology, 7. https://doi.org/10.15626/MP.2019.2058
Rousselet, G. A., & Wilcox, R. R. (2020). Reaction Times and other Skewed Distributions: Problems with the Mean and the Median. Meta-Psychology, 4. https://doi.org/10.15626/MP.2019.1630
Spence, J. R., & Stanley, D. J. (2024). Tempered Expectations: A Tutorial for Calculating and Interpreting Prediction Intervals in the Context of Replications. Advances in Methods and Practices in Psychological Science, 7(1), 25152459231217932. https://doi.org/10.1177/25152459231217932
Verhagen, J., & Wagenmakers, E.-J. (2014). Bayesian tests to quantify the result of a replication attempt. Journal of Experimental Psychology. General, 143(4), 1457–1475. https://doi.org/10.1037/a0036731
Wilcox, R. R., & Rousselet, G. A. (2023). An Updated Guide to Robust Statistical Methods in Neuroscience. Current Protocols, 3(3), e719. https://doi.org/10.1002/cpz1.719
Yan, Y., & Genton, M. G. (2019). The Tukey g-and-h distribution. Significance, 16(3), 12–13. https://doi.org/10.1111/j.1740-9713.2019.01273.x