The R code for this post is on github.
Trimmed means are robust estimators of central tendency. To compute a trimmed mean, we remove a predetermined amount of observations on each side of a distribution, and average the remaining observations. If you think you’re not familiar with trimmed means, you already know one famous member of this family: the median. Indeed, the median is an extreme trimmed mean, in which all observations are removed except one or two.
Using trimmed means confers two advantages:
- trimmed means provide a better estimation of the location of the bulk of the observations than the mean when sampling from asymmetric distributions;
- the standard error of the trimmed mean is less affected by outliers and asymmetry than the mean, so that tests using trimmed means can have more power than tests using the mean.
Important point: if we use a trimmed mean in an inferential test (see below), we make inferences about the population trimmed mean, not the population mean. The same is true for the median or any other measure of central tendency. So each robust estimator is a tool to answer a specific question, and this is why different estimators can return different answers…
Here is how we compute a 20% trimmed mean.
Let’s consider a sample of 20 observations:
39 92 75 61 45 87 59 51 87 12 8 93 74 16 32 39 87 12 47 50
First we sort them:
8 12 12 16 32 39 39 45 47 50 51 59 61 74 75 87 87 87 92 93
The number of observations to remove is floor(0.2 * 20) = 4. So we trim 4 observations from each end:
(8 12 12 16) 32 39 39 45 47 50 51 59 61 74 75 87 (87 87 92 93)
And we take the mean of the remaining observations, such that our 20% trimmed mean = mean(c(32,39,39,45,47,50,51,59,61,74,75,87)) = 54.92
Let’s illustrate the trimming process with a normal distribution and 20% trimming:

We can see how trimming gets rid of the tails of the distribution, to focus on the bulk of the observations. This behaviour is particularly useful when dealing with skewed distributions, as shown here:

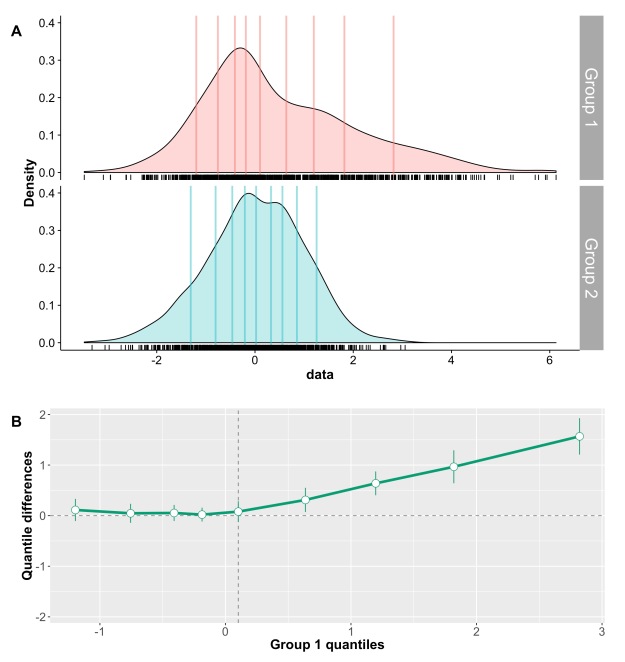
In this skewed distribution (it’s an F distribution), there is more variability on the right side, which appears as stretched compared to the left side. Because we trim the same amount on each side, trimming removes a longer chunk of the distribution on the right side than the left side. As a consequence, the mean of the remaining points is more representative of the location of the bulk of the observations. This can be seen in the following examples.

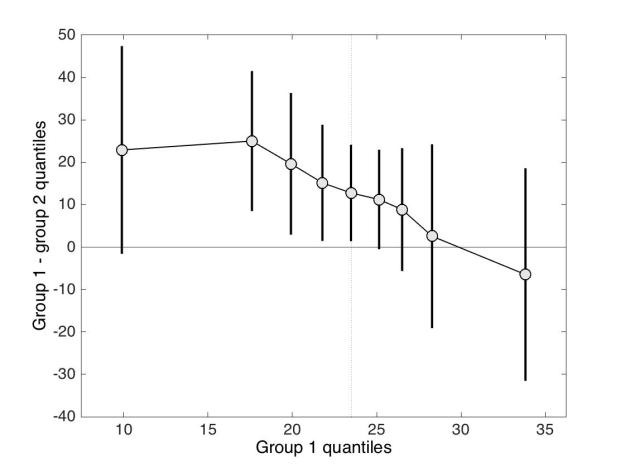
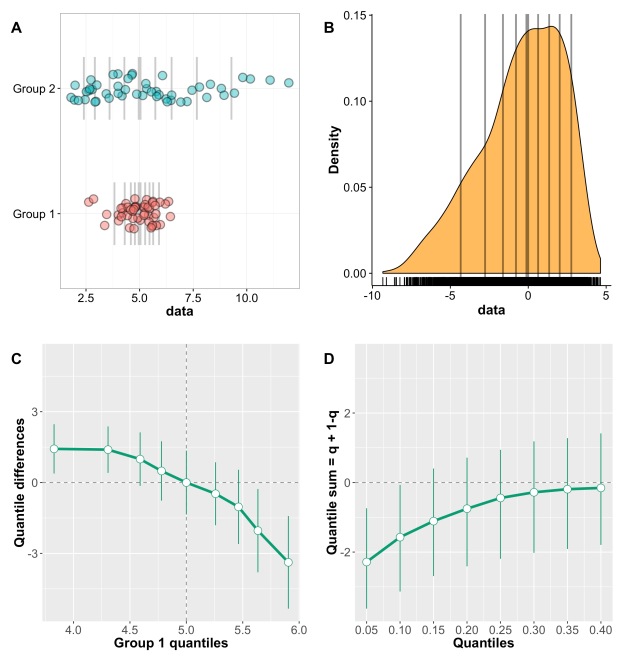
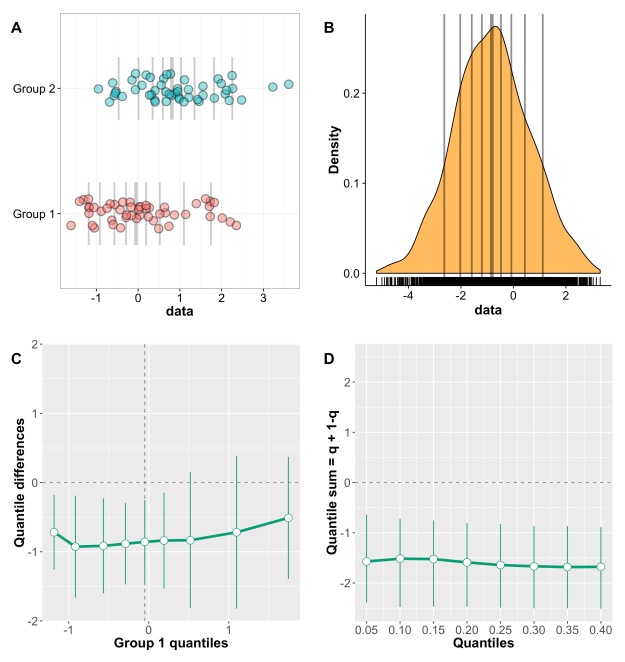
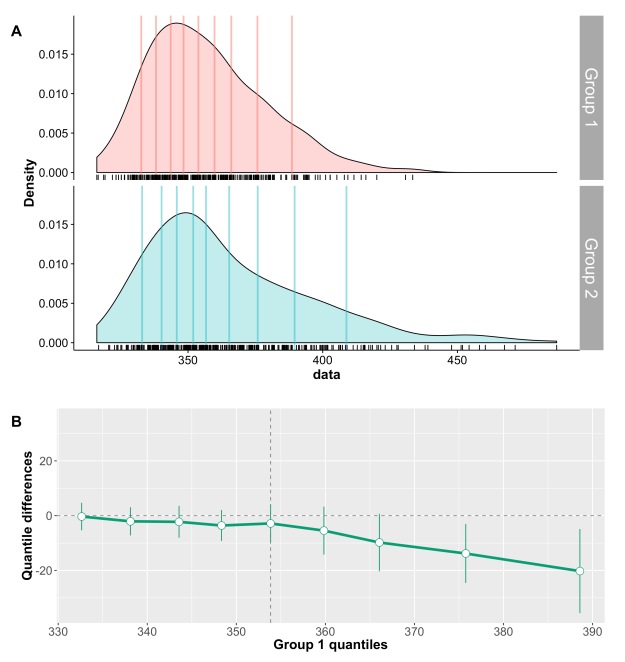
Panel A shows the kernel density estimate of 100 observations sampled from a standard normal distribution (MCT stands for measure of central tendency). By chance, the distribution is not perfectly symmetric, but the mean, 20% trimmed mean and median give very similar estimates, as expected. In panel B, however, the sample is from a lognormal distribution. Because of the asymmetry of the distribution, the mean is dragged towards the right side of the distribution, away from the bulk of the observations. The 20% trimmed mean is to the left of the mean, and the median further to the left, closer to the location of most observations. Thus, for asymmetric distributions, trimmed means provide more accurate information about central tendency than the mean.
**Q: “By trimming, don’t we loose information?”**
I have heard that question over and over. The answer depends on your goal. Statistical methods are only tools to answer specific questions, so it always depends on your goal. I have never met anyone with a true interest in the mean: the mean is always used, implicitly or explicitly, as a tool to indicate the location of the bulk of the observations. Thus, if your goal is to estimate central tendency, then no, trimming doesn’t discard information, it actually increases the quality of the information about central tendency.
I have also heard that criticism: “I’m interested in the tails of the distributions and that’s why I use the mean, trimming gets rid of them”. Tails certainly have interesting stories to tell, but the mean is absolutely not the tool to study them because it mingles all observations into one value, so we have no way to tell why means differ among samples. If you want to study entire distributions, they are fantastic graphical tools available (Rousselet, Pernet & Wilcox 2017).
Implementation
Base R has trimmed means built in:
mean can be used by changing the trim argument to the desired amount of trimming:
mean(x, trim = 0.2) gives a 20% trimmed mean.
In Matlab, try the tm function available here.
In Python, try the scipy.stats.tmean function. More Python functions are listed here.
Inferences
There are plenty of R functions using trimmed means on Rand Wilcox’s website.
We can use trimmed means instead of means in t-tests. However, the calculation of the standard error is different from the traditional t-test formula. This is because after trimming observations, the remaining observations are no longer independent. The formula for the adjusted standard error was originally proposed by Karen Yuen in 1974, and it involves winsorization. To winsorize a sample, instead of removing observations, we replace them with the remaining extreme values. So in our example, a 20% winsorized sample is:
32 32 32 32 32 39 39 45 47 50 51 59 61 74 75 87 87 87 87 87
Taking the mean of the winsorized sample gives a winsorized mean; taking the variance of the winsorized sample gives a winsorized variance etc. I’ve never seen anyone using winsorized means, however the winsorized variance is used to compute the standard error of the trimmed mean (Yuen 1974). There is also a full mathematical explanation in Wilcox (2012).
You can use all the functions below to make inferences about means too, by setting tr=0. How much trimming to use is an empirical question, depending on the type of distributions you deal with. By default, all functions set tr=0.2, 20% trimming, which has been studied a lot and seems to provide a good compromise. Most functions will return an error with an alternative function suggestion if you set tr=0.5: the standard error calculation is inaccurate for the median and often the only satisfactory solution is to use a percentile bootstrap.
**Q: “With trimmed means, isn’t there a danger of users trying different amounts of trimming and reporting the one that give them significant results?”**
This is indeed a possibility, but dishonesty is a property of the user, not a property of the tool. In fact, trying different amounts of trimming could be very informative about the nature of the effects. Reporting the different results, along with graphical representations, could help provide a more detailed description of the effects.
The Yuen t-test performs better than the t-test on means in many situations. For even better results, Wilcox recommends to use trimmed means with a percentile-t bootstrap or a percentile bootstrap. With small amounts of trimming, the percentile-t bootstrap performs better; with at least 20% trimming, the percentile bootstrap is preferable. Details about these choices are available for instance in Wilcox (2012) and Wilcox & Rousselet (2017).
Yuen’s approach
1-alpha confidence interval for the trimmed mean: trimci(x,tr=.2,alpha=0.05)
Yuen t-test for 2 independent groups: yuen(x,y,tr=.2)
Yuen t-test for 2 dependent groups: yuend(x,y,tr=.2)
Bootstrap percentile-t method
One group: trimcibt(x,tr=.2,alpha=.05,nboot=599)
Two independent groups: yuenbt(x,y,tr=.2,alpha=.05,nboot=599)
Two dependent groups: ydbt(x,y,tr=.2,alpha=.05,nboot=599)
Percentile bootstrap approach
One group: trimpb(x,tr=.2,alpha=.05,nboot=2000)
Two independent groups: trimpb2(x,y,tr=.2,alpha=.05,nboot=2000)
Two dependent groups: dtrimpb(x,y=NULL,alpha=.05,con=0,est=mean)
Matlab
There are some Matlab functions here:
tm – trimmed mean
yuen – t-test for 2 independent groups
yuend – t-test for 2 dependent groups
winvar – winsorized variance
winsample – winsorized sample
wincov – winsorized covariance
These functions can be used with several estimators including trimmed means:
pb2dg – percentile bootstrap for 2 dependent groups
pb2ig– percentile bootstrap for 2 independent groups
pbci– percentile bootstrap for 1 group
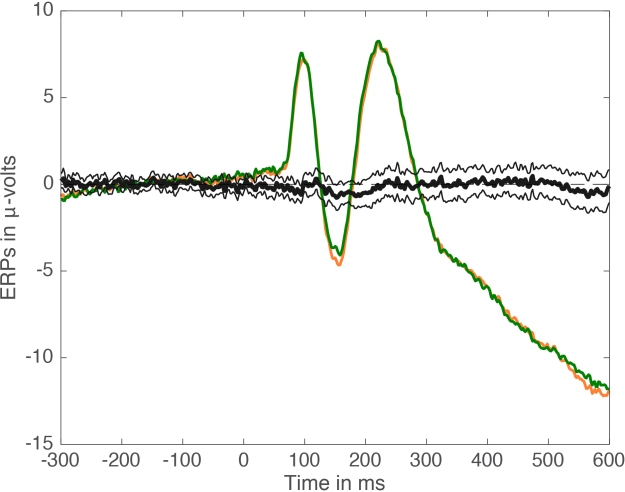
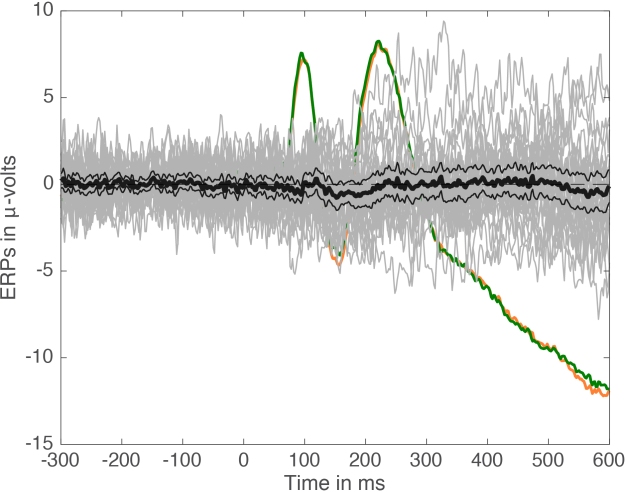
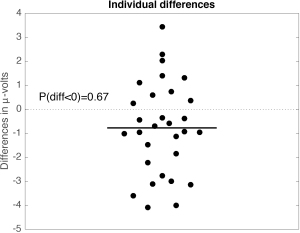
Several functions for trimming large arrays and computing confidence intervals are available in the LIMO EEG toolbox.
References
Karen K. Yuen. The two-sample trimmed t for unequal population variances, Biometrika, Volume 61, Issue 1, 1 April 1974, Pages 165–170, https://doi.org/10.1093/biomet/61.1.165
Rousselet, Guillaume; Pernet, Cyril; Wilcox, Rand (2017): Beyond differences in means: robust graphical methods to compare two groups in neuroscience. figshare. https://doi.org/10.6084/m9.figshare.4055970.v7
Rand R. Wilcox, Guillaume A. Rousselet. A guide to robust statistical methods in neuroscience bioRxiv 151811; doi: https://doi.org/10.1101/151811
Wilcox, R.R. (2012) Introduction to robust estimation and hypothesis testing. Academic Press, San Diego, CA.